|
|
Analogs
-
1529230
-
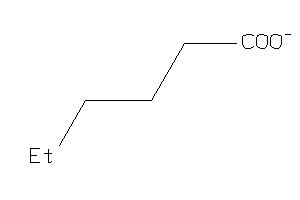
Draw
Identity
99%
90%
80%
70%
Vendors
And 69 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S22A6-2-E |
Solute Carrier Family 22 Member 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
410 |
0.89 |
Binding ≤ 10μM
|
|
Z80583-4-O |
Vero (Kidney Cells) (cluster #4 Of 7), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.36 |
5.1 |
-96.25 |
0 |
4 |
-2 |
80 |
144.126 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.36 |
3.13 |
-45.87 |
1 |
4 |
-1 |
77 |
145.134 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 41 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S22A6-2-E |
Solute Carrier Family 22 Member 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6761 |
0.80 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.15 |
2.5 |
-104.02 |
0 |
4 |
-2 |
80 |
130.099 |
4 |
↓
|
|
|
|
Analogs
-
25762856
-
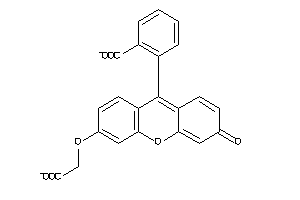
Draw
Identity
99%
90%
80%
70%
Vendors
And 37 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S22A6-3-E |
Solute Carrier Family 22 Member 6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
7.78 |
-58.45 |
1 |
5 |
-1 |
91 |
331.303 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.92 |
7.69 |
-104.25 |
0 |
5 |
-2 |
93 |
330.295 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANM1-1-E |
Protein-arginine N-methyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.28 |
Binding ≤ 10μM
|
|
S22A6-3-E |
Solute Carrier Family 22 Member 6 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
90 |
0.34 |
Binding ≤ 10μM
|
|
S22AK-2-E |
Solute Carrier Family 22 Member 20 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.26 |
Binding ≤ 10μM
|
|
Q5VLE3-2-F |
RmtA (cluster #2 Of 2), Fungal |
Fungi |
180 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.13 |
10.39 |
-94.2 |
0 |
5 |
-2 |
93 |
645.879 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
7.13 |
10.41 |
-51.76 |
1 |
5 |
-1 |
91 |
646.887 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 52 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S22A6-1-E |
Solute Carrier Family 22 Member 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9400 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.46 |
1.51 |
-45.25 |
3 |
5 |
-1 |
95 |
193.182 |
3 |
↓
|
|
|
|
Analogs
-
5760137
-
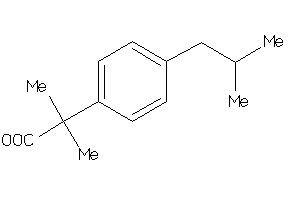
-
5760156
-
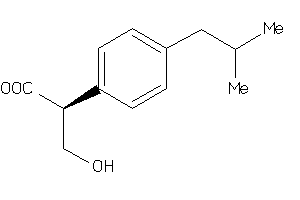
-
5760166
-
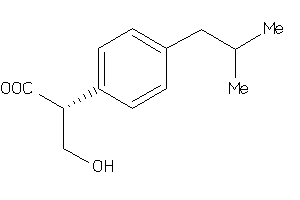
-
2647
-
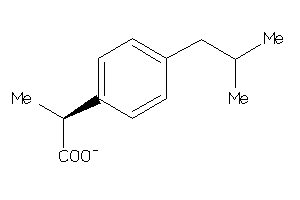
Draw
Identity
99%
90%
80%
70%
Vendors
And 77 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ALBU-1-E |
Serum Albumin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.52 |
Binding ≤ 10μM
|
|
IL8-1-E |
Interleukin-8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.68 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
2000 |
0.53 |
Binding ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1100 |
0.56 |
Binding ≤ 10μM |
|
S22A6-1-E |
Solute Carrier Family 22 Member 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4700 |
0.50 |
Binding ≤ 10μM
|
|
S22AK-1-E |
Solute Carrier Family 22 Member 20 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1122 |
0.56 |
Binding ≤ 10μM
|
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
110 |
0.65 |
Functional ≤ 10μM
|
|
CXCR2-1-E |
C-X-C Chemokine Receptor Type 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
110 |
0.65 |
Functional ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.62 |
Functional ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.46 |
9.02 |
-45.75 |
0 |
2 |
-1 |
40 |
205.277 |
4 |
↓
|
|
|
|
Analogs
-
5760137
-

-
5760156
-

-
5760166
-

-
2647
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 94 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ALBU-1-E |
Serum Albumin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.52 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
2000 |
0.53 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
2600 |
0.52 |
Binding ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1100 |
0.56 |
Binding ≤ 10μM |
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1500 |
0.54 |
Binding ≤ 10μM
|
|
S22A6-1-E |
Solute Carrier Family 22 Member 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4700 |
0.50 |
Binding ≤ 10μM
|
|
S22AK-1-E |
Solute Carrier Family 22 Member 20 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1122 |
0.56 |
Binding ≤ 10μM
|
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.65 |
Functional ≤ 10μM
|
|
CXCR2-1-E |
C-X-C Chemokine Receptor Type 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.65 |
Functional ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.62 |
Functional ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.46 |
9.06 |
-46.94 |
0 |
2 |
-1 |
40 |
205.277 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
5298961
-
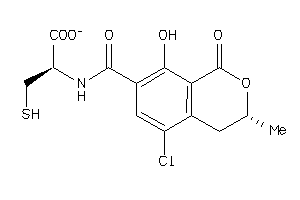
-
5763920
-
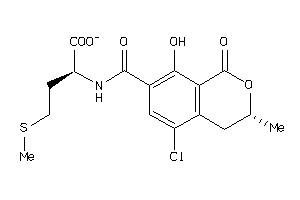
-
25761442
-
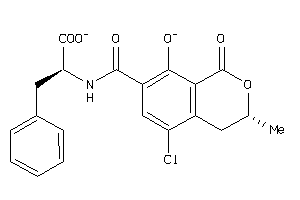
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S22A6-1-E |
Solute Carrier Family 22 Member 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6310 |
0.26 |
Binding ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
60 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.74 |
9.64 |
-59.12 |
2 |
7 |
-1 |
116 |
402.81 |
5 |
↓
|
|
|
|
Analogs
-
37094556
-
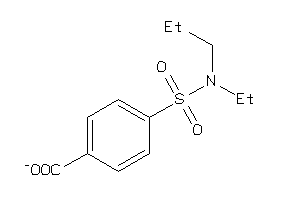
Draw
Identity
99%
90%
80%
70%
Vendors
And 44 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S22A6-1-E |
Solute Carrier Family 22 Member 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6400 |
0.38 |
Binding ≤ 10μM
|
|
S22AK-2-E |
Solute Carrier Family 22 Member 20 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8511 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
6.29 |
-47.89 |
0 |
5 |
-1 |
78 |
284.357 |
7 |
↓
|
|