|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
E2AK2-1-E |
Interferon-induced, Double-stranded RNA-activated Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.52 |
Binding ≤ 10μM |
|
GSK3B-4-E |
Glycogen Synthase Kinase-3 Beta (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
1000 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
1.62 |
-9.06 |
3 |
5 |
0 |
80 |
185.19 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB1-2-E |
G2/mitotic-specific Cyclin B1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.49 |
Binding ≤ 10μM
|
|
CCNB2-2-E |
G2/mitotic-specific Cyclin B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.49 |
Binding ≤ 10μM
|
|
CCNB3-2-E |
G2/mitotic-specific Cyclin B3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.49 |
Binding ≤ 10μM
|
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5000 |
0.49 |
Binding ≤ 10μM
|
|
GSK3B-4-E |
Glycogen Synthase Kinase-3 Beta (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
2300 |
0.53 |
Binding ≤ 10μM
|
|
Z104294-1-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #1 Of 2), Other |
Other |
4000 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
-1.21 |
-8.62 |
1 |
3 |
0 |
41 |
195.225 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA2-3-E |
Cyclin A2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
80 |
0.35 |
Binding ≤ 10μM
|
|
CDK4-1-E |
Cyclin-dependent Kinase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1600 |
0.29 |
Binding ≤ 10μM
|
|
CHK1-1-E |
Serine/threonine-protein Kinase Chk1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.32 |
Binding ≤ 10μM
|
|
DYR1A-3-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
900 |
0.30 |
Binding ≤ 10μM
|
|
GSK3B-4-E |
Glycogen Synthase Kinase-3 Beta (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
40 |
0.37 |
Binding ≤ 10μM
|
|
PDK1-1-E |
Pyruvate Dehydrogenase Kinase Isoform 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.30 |
Binding ≤ 10μM
|
|
PDPK1-1-E |
3-phosphoinositide Dependent Protein Kinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.32 |
Binding ≤ 10μM
|
|
ROCK2-1-E |
Rho-associated Protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.30 |
5.84 |
-17.28 |
4 |
9 |
0 |
136 |
402.48 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.30 |
6.38 |
-43.75 |
5 |
9 |
1 |
137 |
403.488 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.04 |
5.48 |
-18.48 |
4 |
9 |
0 |
139 |
402.48 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKA-3-E |
Serine/threonine-protein Kinase Aurora-A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
58 |
0.44 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
24 |
0.46 |
Binding ≤ 10μM
|
|
GSK3B-4-E |
Glycogen Synthase Kinase-3 Beta (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
24 |
0.46 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
81 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.50 |
9.95 |
-12.37 |
2 |
5 |
0 |
66 |
301.353 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.50 |
10.31 |
-27.16 |
3 |
5 |
1 |
68 |
302.361 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.50 |
10.28 |
-29.66 |
3 |
5 |
1 |
68 |
302.361 |
3 |
↓
|
|
|
|
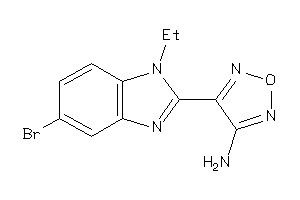
Analogs
-
34372266
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GSK3B-4-E |
Glycogen Synthase Kinase-3 Beta (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
700 |
0.51 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
960 |
0.50 |
Binding ≤ 10μM
|
|
KS6A5-1-E |
Ribosomal Protein S6 Kinase Alpha 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
140 |
0.56 |
Binding ≤ 10μM
|
|
KS6B1-2-E |
Ribosomal Protein S6 Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.60 |
Binding ≤ 10μM
|
|
ROCK1-1-E |
Rho-associated Protein Kinase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
330 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.93 |
5.57 |
-6.64 |
2 |
6 |
0 |
83 |
229.243 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2345 |
0.53 |
Binding ≤ 10μM
|
|
GSK3B-4-E |
Glycogen Synthase Kinase-3 Beta (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
2343 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.47 |
4.12 |
-16.04 |
2 |
5 |
0 |
71 |
204.233 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
530 |
0.55 |
Binding ≤ 10μM
|
|
GSK3B-4-E |
Glycogen Synthase Kinase-3 Beta (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
530 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.69 |
3.88 |
-18.61 |
3 |
5 |
0 |
80 |
211.228 |
1 |
↓
|
|
Ref
Reference (pH 7)
|
1.69 |
3.92 |
-10.61 |
3 |
5 |
0 |
80 |
211.228 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM
|
|
CHK1-1-E |
Serine/threonine-protein Kinase Chk1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.42 |
Binding ≤ 10μM
|
|
CHK2-1-E |
Serine/threonine-protein Kinase Chk2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.37 |
Binding ≤ 10μM
|
|
E2AK2-1-E |
Interferon-induced, Double-stranded RNA-activated Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM |
|
GSK3B-4-E |
Glycogen Synthase Kinase-3 Beta (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
500 |
0.42 |
Binding ≤ 10μM
|
|
PRKDC-1-E |
DNA-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
10000 |
0.33 |
Functional ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
1000 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.55 |
5.93 |
-10.93 |
2 |
6 |
0 |
83 |
276.255 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.55 |
6.42 |
-44.52 |
3 |
6 |
1 |
84 |
277.263 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GSK3B-4-E |
Glycogen Synthase Kinase-3 Beta (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
7300 |
0.45 |
Binding ≤ 10μM
|
|
KS6B1-2-E |
Ribosomal Protein S6 Kinase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
540 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.56 |
4.27 |
-7.75 |
2 |
6 |
0 |
83 |
215.216 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.56 |
4.1 |
-30.52 |
3 |
6 |
1 |
84 |
216.224 |
1 |
↓
|
|
|
|
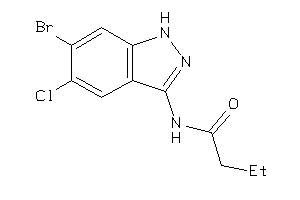
Analogs
-
38781931
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GSK3B-4-E |
Glycogen Synthase Kinase-3 Beta (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
198 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
5.48 |
-12.61 |
2 |
4 |
0 |
58 |
282.141 |
3 |
↓
|
|