|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRXR2-1-E |
Thioredoxin Reductase 2, Mitochondrial (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.42 |
Binding ≤ 10μM
|
|
Z50459-5-O |
Leishmania Donovani (cluster #5 Of 8), Other |
Other |
1300 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
7.16 |
-5.16 |
0 |
6 |
0 |
92 |
270.55 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRXR1-3-E |
Thioredoxin Reductase 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.30 |
Binding ≤ 10μM
|
|
TRXR2-1-E |
Thioredoxin Reductase 2, Mitochondrial (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.30 |
Binding ≤ 10μM
|
|
TRXR3-1-E |
Thioredoxin Reductase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.60 |
11.05 |
-14.05 |
0 |
12 |
0 |
183 |
366.267 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRXR1-3-E |
Thioredoxin Reductase 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.53 |
Binding ≤ 10μM
|
|
TRXR2-1-E |
Thioredoxin Reductase 2, Mitochondrial (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.53 |
Binding ≤ 10μM
|
|
TRXR3-1-E |
Thioredoxin Reductase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.86 |
4 |
-8.26 |
0 |
8 |
0 |
117 |
226.173 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRXR2-1-E |
Thioredoxin Reductase 2, Mitochondrial (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
3.22 |
-5.92 |
0 |
5 |
0 |
72 |
181.176 |
1 |
↓
|
|
|
|
Analogs
-
39240406
-
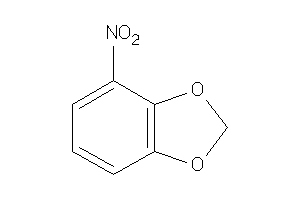
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRXR2-1-E |
Thioredoxin Reductase 2, Mitochondrial (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.67 |
5.2 |
-16.65 |
0 |
8 |
0 |
110 |
212.117 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRXR2-1-E |
Thioredoxin Reductase 2, Mitochondrial (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.56 |
4.69 |
-13.61 |
0 |
8 |
0 |
117 |
220.144 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 40 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRXR2-1-E |
Thioredoxin Reductase 2, Mitochondrial (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
650 |
0.72 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
2900 |
0.65 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.94 |
3.35 |
-12.83 |
0 |
5 |
0 |
72 |
181.176 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRXR1-1-E |
Thioredoxin Reductase 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8100 |
0.18 |
Binding ≤ 10μM
|
|
TRXR2-1-E |
Thioredoxin Reductase 2, Mitochondrial (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7900 |
0.18 |
Binding ≤ 10μM
|
|
GSHR-1-F |
Glutathione Reductase (cluster #1 Of 1), Fungal |
Fungi |
2200 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.55 |
-9.06 |
-20.41 |
7 |
9 |
0 |
175 |
520.449 |
1 |
↓
|
|