|
|
Analogs
-
1531008
-
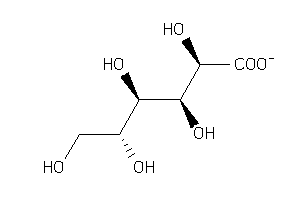
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,3S,4S,5R)-2,3,4,5-Tetrahydroxy-6-oxohexanoic acid
(2S,3S,4S,5R)-2,3,4,5-Tetrahydro…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.77 |
-5.54 |
-49.93 |
4 |
7 |
-1 |
138 |
193.131 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
-2.77 |
-5.47 |
-49.2 |
4 |
7 |
-1 |
138 |
193.131 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
-2.77 |
-4.49 |
-49.62 |
4 |
7 |
-1 |
138 |
193.131 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Mid
Mid (pH 6-8)
|
-2.71 |
2.08 |
-61.33 |
2 |
6 |
-1 |
98 |
182.159 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.21 |
0.16 |
-65.58 |
3 |
6 |
0 |
103 |
197.194 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.73 |
2.82 |
-62.13 |
2 |
6 |
-1 |
98 |
196.186 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.21 |
1.7 |
-82.21 |
4 |
6 |
1 |
104 |
198.202 |
4 |
↓
|
|
|
|
Analogs
-
13219479
-
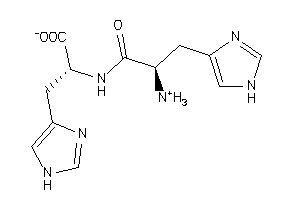
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.88 |
2.66 |
-101.42 |
7 |
9 |
1 |
155 |
293.307 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.88 |
1.89 |
-63 |
5 |
9 |
-1 |
153 |
291.291 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.88 |
2.18 |
-62.13 |
6 |
9 |
0 |
154 |
292.299 |
7 |
↓
|
|
|
|
Analogs
-
38200491
-
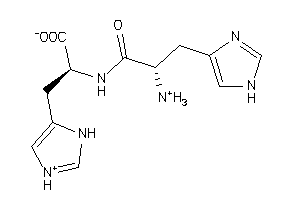
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.88 |
2.76 |
-101.6 |
7 |
9 |
1 |
155 |
293.307 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.88 |
2.36 |
-55.69 |
5 |
9 |
-1 |
153 |
291.291 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.88 |
1.93 |
-52.41 |
5 |
9 |
-1 |
153 |
291.291 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.38 |
-1.39 |
-227.43 |
1 |
7 |
-3 |
133 |
183.032 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.38 |
-2.53 |
-113.57 |
2 |
7 |
-2 |
130 |
184.04 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.38 |
-1.38 |
-227.49 |
1 |
7 |
-3 |
133 |
183.032 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.38 |
-2.53 |
-114.1 |
2 |
7 |
-2 |
130 |
184.04 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PEPD-1-E |
Xaa-Pro Dipeptidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9 |
1.13 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.80 |
1.7 |
-214.18 |
0 |
6 |
-3 |
113 |
165.017 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.80 |
0.53 |
-102.67 |
1 |
6 |
-2 |
110 |
166.025 |
3 |
↓
|
|
|
|
Analogs
-
1532655
-
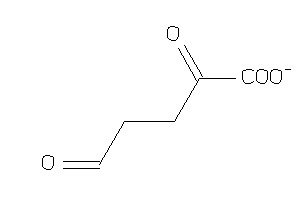
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGLN1-2-E |
Egl Nine Homolog 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.85 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.48 |
4.22 |
-95.01 |
0 |
5 |
-2 |
97 |
144.082 |
4 |
↓
|
|
|
|
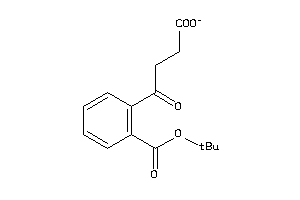
Analogs
-
2579857
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.77 |
0.99 |
-119.22 |
0 |
5 |
-2 |
97 |
220.18 |
5 |
↓
|
|
|
|
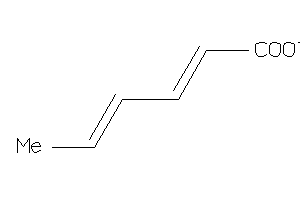
Analogs
-
1558385
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.16 |
2.58 |
-112.43 |
0 |
4 |
-2 |
80 |
140.094 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.16 |
1.48 |
-55.15 |
1 |
4 |
-1 |
77 |
141.102 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
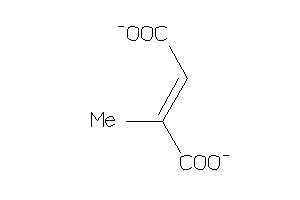
Analogs
-
4501410
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.13 |
2.55 |
-127.37 |
0 |
4 |
-2 |
80 |
128.083 |
2 |
↓
|
|
|
|
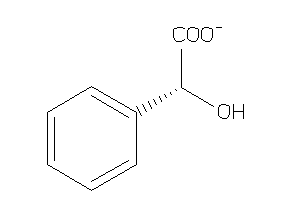
Analogs
-
164392
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.37 |
3.51 |
-45.3 |
1 |
3 |
-1 |
60 |
151.141 |
2 |
↓
|
|
|
|
Analogs
-
36
-
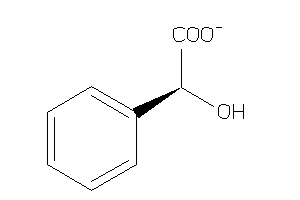
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.37 |
3.53 |
-45.2 |
1 |
3 |
-1 |
60 |
151.141 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.04 |
-2.25 |
-55.9 |
3 |
5 |
-1 |
107 |
146.122 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.04 |
-2.29 |
-54.31 |
3 |
5 |
-1 |
107 |
146.122 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.04 |
-2.29 |
-54.28 |
3 |
5 |
-1 |
107 |
146.122 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.04 |
-2.27 |
-58.17 |
3 |
5 |
-1 |
107 |
146.122 |
3 |
↓
|
|
|
|
Analogs
-
1531008
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.77 |
-6.69 |
-51.79 |
4 |
7 |
-1 |
138 |
193.131 |
5 |
↓
|
|
|
|
Analogs
-
901090
-
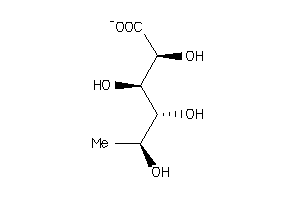
-
1531008
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.76 |
-5.11 |
-50.41 |
4 |
7 |
-1 |
138 |
193.131 |
5 |
↓
|
|
|
|
Analogs
-
1531008
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.76 |
-6.33 |
-50.29 |
4 |
7 |
-1 |
138 |
193.131 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.22 |
-9.7 |
-50.49 |
5 |
7 |
-1 |
141 |
195.147 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.22 |
-10.1 |
-50.89 |
5 |
7 |
-1 |
141 |
195.147 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.22 |
-9.67 |
-50.18 |
5 |
7 |
-1 |
141 |
195.147 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.22 |
-10.91 |
-51.51 |
5 |
7 |
-1 |
141 |
195.147 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.22 |
-10.81 |
-50.68 |
5 |
7 |
-1 |
141 |
195.147 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.22 |
-9.67 |
-50.2 |
5 |
7 |
-1 |
141 |
195.147 |
5 |
↓
|
|
|
|
Analogs
-
901090
-

-
1531008
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.22 |
-10.1 |
-50.89 |
5 |
7 |
-1 |
141 |
195.147 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.22 |
-9.7 |
-50.48 |
5 |
7 |
-1 |
141 |
195.147 |
5 |
↓
|
|
|
|
Analogs
-
8551175
-
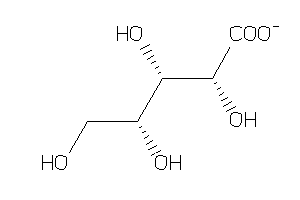
-
8551176
-
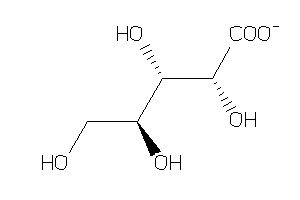
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.79 |
-8.04 |
-47.2 |
4 |
6 |
-1 |
121 |
165.121 |
4 |
↓
|
|
|
|
Analogs
-
8551175
-

-
8551176
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.79 |
-8.37 |
-47.16 |
4 |
6 |
-1 |
121 |
165.121 |
4 |
↓
|
|
|
|
Analogs
-
8551175
-

-
8551176
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.79 |
-7.74 |
-47.92 |
4 |
6 |
-1 |
121 |
165.121 |
4 |
↓
|
|
|
|
Analogs
-
8551175
-

-
8551176
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.79 |
-8.87 |
-47.87 |
4 |
6 |
-1 |
121 |
165.121 |
4 |
↓
|
|
|
|
Analogs
-
8551175
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.79 |
-8.87 |
-47.88 |
4 |
6 |
-1 |
121 |
165.121 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
8551175
-

-
8551176
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.79 |
-8.04 |
-47.23 |
4 |
6 |
-1 |
121 |
165.121 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.34 |
-4.4 |
-97.37 |
4 |
8 |
-2 |
161 |
208.122 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.34 |
-4.07 |
-97.65 |
4 |
8 |
-2 |
161 |
208.122 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
1531008
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.34 |
-7.36 |
-99.81 |
4 |
8 |
-2 |
161 |
208.122 |
5 |
↓
|
|
|
|
Analogs
-
901090
-

-
1531008
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.34 |
-6.67 |
-100.54 |
4 |
8 |
-2 |
161 |
208.122 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA4-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
530 |
0.63 |
Binding ≤ 10μM
|
|
ACHA6-3-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
530 |
0.63 |
Binding ≤ 10μM
|
|
ACHA7-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
5770 |
0.52 |
Binding ≤ 10μM
|
|
ACHB3-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
530 |
0.63 |
Binding ≤ 10μM
|
|
ACHB4-7-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
24 |
0.76 |
Binding ≤ 10μM
|
|
Q8BGP7-2-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
530 |
0.63 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA4_MOUSE |
O70174
|
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit, Mouse |
530 |
0.63 |
Binding ≤ 1μM
|
|
Q8BGP7_MOUSE |
Q8BGP7
|
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit, Mouse |
30 |
0.75 |
Binding ≤ 1μM
|
|
ACHA6_MOUSE |
Q9R0W9
|
Neuronal Acetylcholine Receptor Subunit Alpha-6, Mouse |
340 |
0.65 |
Binding ≤ 1μM
|
|
ACHB3_MOUSE |
Q8BMN3
|
Neuronal Acetylcholine Receptor Subunit Beta-3, Mouse |
530 |
0.63 |
Binding ≤ 1μM
|
|
ACHB4_MOUSE |
Q8R493
|
Neuronal Acetylcholine Receptor Subunit Beta-4, Mouse |
30 |
0.75 |
Binding ≤ 1μM
|
|
ACHA4_MOUSE |
O70174
|
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit, Mouse |
1200 |
0.59 |
Binding ≤ 10μM
|
|
ACHA7_MOUSE |
P49582
|
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit, Mouse |
5770 |
0.52 |
Binding ≤ 10μM
|
|
Q8BGP7_MOUSE |
Q8BGP7
|
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit, Mouse |
1200 |
0.59 |
Binding ≤ 10μM
|
|
ACHA6_MOUSE |
Q9R0W9
|
Neuronal Acetylcholine Receptor Subunit Alpha-6, Mouse |
1200 |
0.59 |
Binding ≤ 10μM
|
|
ACHB3_MOUSE |
Q8BMN3
|
Neuronal Acetylcholine Receptor Subunit Beta-3, Mouse |
1200 |
0.59 |
Binding ≤ 10μM
|
|
ACHB4_MOUSE |
Q8R493
|
Neuronal Acetylcholine Receptor Subunit Beta-4, Mouse |
30 |
0.75 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.34 |
-8.52 |
-100.41 |
4 |
8 |
-2 |
161 |
208.122 |
5 |
↓
|
|
|
|
Analogs
-
901090
-

-
1531008
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.34 |
-3.44 |
-97.28 |
4 |
8 |
-2 |
161 |
208.122 |
5 |
↓
|
|
|
|
Analogs
-
901090
-

-
1531008
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.34 |
-7.69 |
-99.91 |
4 |
8 |
-2 |
161 |
208.122 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.34 |
-5.32 |
-99.81 |
4 |
8 |
-2 |
161 |
208.122 |
5 |
↓
|
|