|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.59 |
-3.5 |
-11.32 |
2 |
9 |
0 |
156 |
254.087 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
-1.93 |
-6.93 |
-238.98 |
3 |
9 |
-3 |
169 |
253.079 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.93 |
-8.07 |
-117.79 |
4 |
9 |
-2 |
166 |
254.087 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-5-O |
Plasmodium Falciparum (cluster #5 Of 22), Other |
Other |
10000 |
0.88 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.88 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.70 |
-0.64 |
-115.99 |
1 |
5 |
-2 |
100 |
138.015 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.70 |
0.47 |
-223.24 |
0 |
5 |
-3 |
103 |
137.007 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
8215517
-
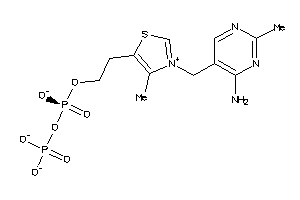
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.77 |
-3.12 |
-135.84 |
2 |
6 |
-2 |
113 |
168.041 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
-2.45 |
-1.03 |
-138.28 |
1 |
6 |
-2 |
110 |
168.041 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
-1.81 |
-1.29 |
-130.45 |
1 |
6 |
-2 |
110 |
168.041 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
|
|
|
Analogs
-
12402869
-
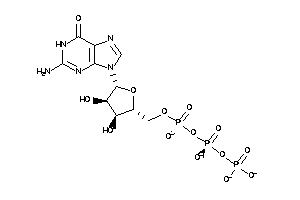
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.22 |
-7.52 |
-180.6 |
6 |
19 |
-3 |
308 |
520.157 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.53 |
-4.25 |
-237.08 |
6 |
19 |
-3 |
308 |
520.157 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
61 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPR17-1-E |
Uracil Nucleotide/cysteinyl Leukotriene Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1140 |
0.33 |
Binding ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
15 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
130 |
0.33 |
Functional ≤ 10μM
|
|
P2RY4-1-E |
Pyrimidinergic Receptor P2Y4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
90 |
0.34 |
Functional ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5NTD-1-E |
5'-nucleotidase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.47 |
Binding ≤ 10μM
|
|
GRP78-1-E |
78 KDa Glucose-regulated Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3830 |
0.28 |
Binding ≤ 10μM
|
|
HSP7C-1-E |
Heat Shock Cognate 71 KDa Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.36 |
Binding ≤ 10μM
|
|
KAP2-1-E |
CAMP-dependent Protein Kinase Type II-alpha Regulatory Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.36 |
Functional ≤ 10μM
|
|
P2Y12-1-E |
Purinergic Receptor P2Y12 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
69 |
0.37 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5NTD_RAT |
P21588
|
5'-nucleotidase, Rat |
0.91 |
0.47 |
Binding ≤ 1μM
|
|
HSP7C_BOVIN |
P19120
|
Heat Shock Cognate 71 KDa Protein, Bovin |
110 |
0.36 |
Binding ≤ 1μM
|
|
HSP7C_HUMAN |
P11142
|
Heat Shock Cognate 71 KDa Protein, Human |
260 |
0.34 |
Binding ≤ 1μM
|
|
5NTD_RAT |
P21588
|
5'-nucleotidase, Rat |
0.91 |
0.47 |
Binding ≤ 10μM
|
|
GRP78_HUMAN |
P11021
|
78 KDa Glucose-regulated Protein, Human |
2170 |
0.29 |
Binding ≤ 10μM
|
|
KAPCA_MOUSE |
P05132
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Mouse |
10000 |
0.26 |
Binding ≤ 10μM
|
|
KAPCB_MOUSE |
P68181
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Mouse |
10000 |
0.26 |
Binding ≤ 10μM
|
|
KAP2_BOVIN |
P00515
|
CAMP-dependent Protein Kinase Type II-alpha Regulatory Subunit, Bovin |
10000 |
0.26 |
Binding ≤ 10μM
|
|
HSP7C_BOVIN |
P19120
|
Heat Shock Cognate 71 KDa Protein, Bovin |
110 |
0.36 |
Binding ≤ 10μM
|
|
HSP7C_HUMAN |
P11142
|
Heat Shock Cognate 71 KDa Protein, Human |
260 |
0.34 |
Binding ≤ 10μM
|
|
P2RX1_HUMAN |
P51575
|
P2X Purinoceptor 1, Human |
10000 |
0.26 |
Functional ≤ 10μM
|
|
P2RY1_MELGA |
P49652
|
P2Y Purinoceptor 1, Melga |
100 |
0.36 |
Functional ≤ 10μM
|
|
P2RY1_HUMAN |
P47900
|
Purinergic Receptor P2Y1, Human |
14 |
0.41 |
Functional ≤ 10μM
|
|
P2Y12_RAT |
Q9EPX4
|
Purinergic Receptor P2Y12, Rat |
69 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Hi
High (pH 8-9.5)
|
3.79 |
9.28 |
-36.43 |
1 |
2 |
1 |
8 |
233.379 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAP2-1-E |
CAMP-dependent Protein Kinase Type II-alpha Regulatory Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6300 |
0.23 |
Binding ≤ 10μM
|
|
P2RY1-1-E |
Purinergic Receptor P2Y1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.25 |
Binding ≤ 10μM
|
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
56 |
0.33 |
Functional ≤ 10μM
|
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.26 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
340 |
0.29 |
Functional ≤ 10μM
|
|
P2RX4-1-E |
P2X Purinoceptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.28 |
Functional ≤ 10μM
|
|
P2RX5-1-E |
P2X Purinoceptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RX6-1-E |
P2X Purinoceptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.23 |
Functional ≤ 10μM
|
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
96 |
0.32 |
Functional ≤ 10μM
|
|
P2RY4-1-E |
Pyrimidinergic Receptor P2Y4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.26 |
Functional ≤ 10μM
|
|
P2Y11-1-E |
Purinergic Receptor P2Y11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.25 |
Functional ≤ 10μM |
|
P2Y12-1-E |
Purinergic Receptor P2Y12 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3190 |
0.25 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA_MOUSE |
P05132
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Mouse |
10000 |
0.23 |
Binding ≤ 10μM
|
|
KAPCB_MOUSE |
P68181
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Mouse |
10000 |
0.23 |
Binding ≤ 10μM
|
|
KAP2_BOVIN |
P00515
|
CAMP-dependent Protein Kinase Type II-alpha Regulatory Subunit, Bovin |
10000 |
0.23 |
Binding ≤ 10μM
|
|
MAPK2_HUMAN |
P49137
|
MAP Kinase-activated Protein Kinase 2, Human |
2800 |
0.25 |
Binding ≤ 10μM
|
|
P2RY1_HUMAN |
P47900
|
Purinergic Receptor P2Y1, Human |
1500 |
0.26 |
Binding ≤ 10μM
|
|
P2RX1_HUMAN |
P51575
|
P2X Purinoceptor 1, Human |
56 |
0.33 |
Functional ≤ 10μM
|
|
P2RX2_RAT |
P49653
|
P2X Purinoceptor 2, Rat |
1400 |
0.26 |
Functional ≤ 10μM
|
|
P2RX3_HUMAN |
P56373
|
P2X Purinoceptor 3, Human |
340 |
0.29 |
Functional ≤ 10μM
|
|
P2RX4_HUMAN |
Q99571
|
P2X Purinoceptor 4, Human |
500 |
0.28 |
Functional ≤ 10μM
|
|
P2RX4_RAT |
P51577
|
P2X Purinoceptor 4, Rat |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RX5_RAT |
P51578
|
P2X Purinoceptor 5, Rat |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RX6_RAT |
P51579
|
P2X Purinoceptor 6, Rat |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RY1_HUMAN |
P47900
|
Purinergic Receptor P2Y1, Human |
14 |
0.35 |
Functional ≤ 10μM
|
|
P2Y11_HUMAN |
Q96G91
|
Purinergic Receptor P2Y11, Human |
3300 |
0.25 |
Functional ≤ 10μM |
|
P2Y12_RAT |
Q9EPX4
|
Purinergic Receptor P2Y12, Rat |
3190 |
0.25 |
Functional ≤ 10μM
|
|
P2RY2_HUMAN |
P41231
|
Purinergic Receptor P2Y2, Human |
230 |
0.30 |
Functional ≤ 10μM
|
|
P2RY4_RAT |
O35811
|
Pyrimidinergic Receptor P2Y4, Rat |
1800 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.39 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
590 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.93 |
2.3 |
-113.35 |
0 |
5 |
-1 |
72 |
182.136 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.38 |
-1.39 |
-227.43 |
1 |
7 |
-3 |
133 |
183.032 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.38 |
-2.53 |
-113.57 |
2 |
7 |
-2 |
130 |
184.04 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPOL-3-V |
Human Herpesvirus 6 DNA Polymerase (cluster #3 Of 4), Viral |
Viruses |
280 |
1.31 |
Binding ≤ 10μM
|
|
Q72547-6-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #6 Of 6), Viral |
Viruses |
2030 |
1.14 |
Binding ≤ 10μM
|
|
Z50613-1-O |
Avian Myeloblastosis Virus (cluster #1 Of 1), Other |
Other |
9310 |
1.01 |
Binding ≤ 10μM
|
|
Z50530-5-O |
Human Herpesvirus 5 (cluster #5 Of 5), Other |
Other |
7000 |
1.03 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.71 |
-2.5 |
-120.9 |
1 |
5 |
-2 |
100 |
123.988 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.16 |
-1.64 |
-123.1 |
3 |
8 |
-2 |
144 |
209.098 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
4096362
-
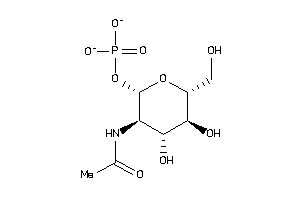
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.75 |
-10.63 |
-151.53 |
4 |
10 |
-2 |
171 |
299.172 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.38 |
-6.71 |
-120.99 |
7 |
9 |
-1 |
180 |
253.175 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.11 |
4.51 |
-121.31 |
2 |
8 |
-1 |
128 |
343.325 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.11 |
3.36 |
-49.82 |
3 |
8 |
0 |
125 |
344.333 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-4.11 |
3.81 |
-98.96 |
4 |
8 |
1 |
127 |
345.341 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.37 |
-6.99 |
-324.8 |
3 |
12 |
-4 |
215 |
336.082 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.37 |
-8.15 |
-209.63 |
4 |
12 |
-3 |
212 |
337.09 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-3.37 |
-9.05 |
-117.34 |
5 |
12 |
-2 |
209 |
338.098 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.76 |
0.89 |
-129.43 |
1 |
7 |
-2 |
123 |
245.127 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.76 |
1.89 |
-195.99 |
0 |
7 |
-3 |
125 |
244.119 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.76 |
-0.27 |
-42.2 |
2 |
7 |
-1 |
120 |
246.135 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PEPD-1-E |
Xaa-Pro Dipeptidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9 |
1.13 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.80 |
1.7 |
-214.18 |
0 |
6 |
-3 |
113 |
165.017 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.80 |
0.53 |
-102.67 |
1 |
6 |
-2 |
110 |
166.025 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-5-O |
Plasmodium Falciparum (cluster #5 Of 22), Other |
Other |
84 |
0.99 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
47.1 |
1.03 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.26 |
-2.26 |
-49.62 |
4 |
6 |
-1 |
128 |
168.065 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-5-O |
Plasmodium Falciparum (cluster #5 Of 22), Other |
Other |
84 |
0.99 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
47.1 |
1.03 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.26 |
-2.32 |
-49.76 |
4 |
6 |
-1 |
128 |
168.065 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AROA-2-B |
5-enolpyruvylshikimate-3-phosphate Synthase (cluster #2 Of 2), Bacterial |
Bacteria |
2000 |
0.80 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.84 |
-0.07 |
-110.02 |
2 |
6 |
-2 |
120 |
167.057 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.84 |
-1.18 |
-47.65 |
3 |
6 |
-1 |
117 |
168.065 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.38 |
2.51 |
-134.92 |
0 |
4 |
-2 |
72 |
300.335 |
9 |
↓
|
|
|
|
Analogs
-
12501438
-
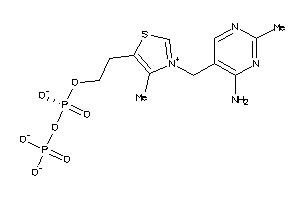
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.75 |
4.38 |
-217.61 |
2 |
11 |
-2 |
177 |
422.296 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.75 |
3.22 |
-112.63 |
3 |
11 |
-1 |
175 |
423.304 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-4.75 |
3.67 |
-154.1 |
4 |
11 |
0 |
176 |
424.312 |
8 |
↓
|
|
|
|
Analogs
-
5785202
-
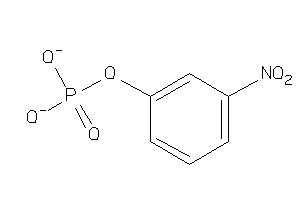
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
330 |
0.65 |
Binding ≤ 10μM
|
|
CAH13-7-E |
Carbonic Anhydrase XIII (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
1050 |
0.60 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
63 |
0.72 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.67 |
4.09 |
-121.92 |
0 |
7 |
-2 |
118 |
217.073 |
3 |
↓
|
|
|
|
|
|
|
Analogs
-
1655964
-
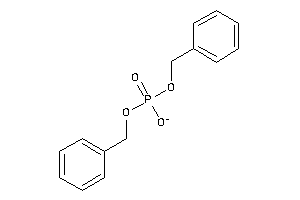
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.61 |
3.8 |
-137.26 |
0 |
4 |
-2 |
72 |
186.103 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.18 |
-3.36 |
-116.8 |
3 |
5 |
-1 |
100 |
140.055 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.81 |
-1.91 |
-130.51 |
1 |
6 |
-2 |
109 |
168.041 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.26 |
-3.31 |
-137.8 |
2 |
6 |
-2 |
113 |
170.057 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.26 |
-4.46 |
-46.79 |
3 |
6 |
-1 |
110 |
171.065 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.38 |
-0.86 |
-203.04 |
1 |
7 |
-3 |
133 |
183.032 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.38 |
-2.02 |
-98.17 |
2 |
7 |
-2 |
130 |
184.04 |
4 |
↓
|
|