|
|
Analogs
-
5390388
-
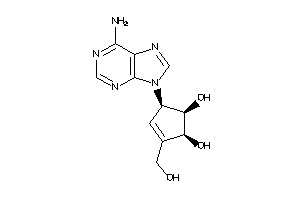
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.31 |
-2.18 |
-17.63 |
5 |
8 |
0 |
130 |
263.257 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
36152036
-

-
36152025
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
200 |
0.32 |
Binding ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
200 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.37 |
3.3 |
-353.33 |
2 |
15 |
-4 |
241 |
467.164 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.37 |
2.14 |
-216.98 |
3 |
15 |
-3 |
238 |
468.172 |
8 |
↓
|
|
|
|
Analogs
-
36152025
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
200 |
0.32 |
Binding ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
200 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.37 |
3.3 |
-349.63 |
2 |
15 |
-4 |
241 |
467.164 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.37 |
2.15 |
-214.59 |
3 |
15 |
-3 |
238 |
468.172 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.48 |
2.42 |
-11.3 |
5 |
7 |
0 |
116 |
246.274 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.48 |
-2.62 |
-28.49 |
6 |
7 |
1 |
117 |
247.282 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.48 |
2.66 |
-28.44 |
6 |
7 |
1 |
117 |
247.282 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.85 |
-1.19 |
-16.84 |
5 |
8 |
0 |
130 |
342.153 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.85 |
-0.44 |
-47.48 |
4 |
8 |
-1 |
133 |
341.145 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.15 |
1.06 |
-14.67 |
4 |
7 |
0 |
110 |
312.127 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.12 |
1.56 |
-13.96 |
4 |
7 |
0 |
110 |
359.127 |
1 |
↓
|
|