|
|
Analogs
-
8382353
-
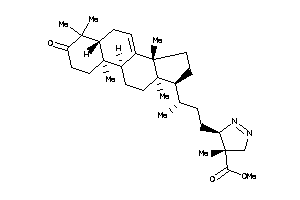
-
8382354
-
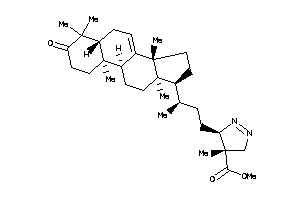
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.88 |
11.93 |
-8.45 |
0 |
5 |
0 |
68 |
510.763 |
6 |
↓
|
|
|
|
Analogs
-
8382353
-

-
8382354
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.88 |
12.1 |
-8.65 |
0 |
5 |
0 |
68 |
510.763 |
6 |
↓
|
|
|
|
Analogs
-
8382353
-

-
8382354
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.88 |
12.13 |
-8.68 |
0 |
5 |
0 |
68 |
510.763 |
6 |
↓
|
|
|
|
Analogs
-
8382353
-

-
8382354
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.88 |
11.96 |
-8.43 |
0 |
5 |
0 |
68 |
510.763 |
6 |
↓
|
|
|
|
Analogs
-
33834014
-
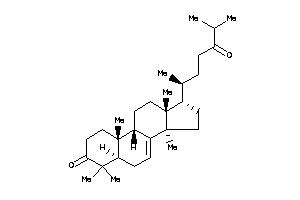
-
5761206
-
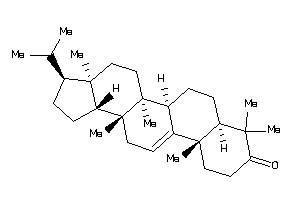
-
5761217
-
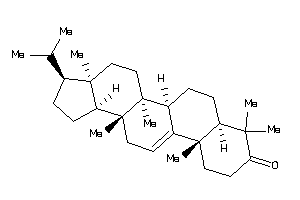
-
5761432
-
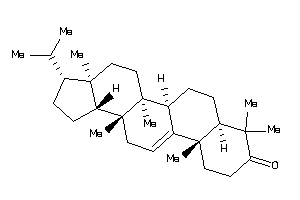
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5S,9S,10R,13S,14S,17S)-17-[(1S)-1,5-dimethylhex-4-enyl]-4,4,10,13,14-pentamethyl-1,2,5,6,9,11,12,15
(5S,9S,10R,13S,14S,17S)-17-[(1S)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.47 |
16.01 |
-4.22 |
0 |
1 |
0 |
17 |
424.713 |
4 |
↓
|
|
|
|
Analogs
-
33834014
-

-
5761206
-

-
5761217
-

-
5761432
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(5S,9S,10R,13S,14S,17S)-17-[(1R)-1,5-dimethylhex-4-enyl]-4,4,10,13,14-pentamethyl-1,2,5,6,9,11,12,15
(5S,9S,10R,13S,14S,17S)-17-[(1R)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.47 |
15.97 |
-3.99 |
0 |
1 |
0 |
17 |
424.713 |
4 |
↓
|
|
|
|
Analogs
-
33834014
-

-
5761206
-

-
5761217
-

-
5761432
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(5S,9S,10R,13S,14S,17R)-17-[(1S)-1,5-dimethylhex-4-enyl]-4,4,10,13,14-pentamethyl-1,2,5,6,9,11,12,15
(5S,9S,10R,13S,14S,17R)-17-[(1S)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.47 |
15.97 |
-4.8 |
0 |
1 |
0 |
17 |
424.713 |
4 |
↓
|
|
|
|
Analogs
-
33834014
-

-
5761206
-

-
5761217
-

-
5761432
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(5S,9S,10R,13S,14S,17R)-17-[(1R)-1,5-dimethylhex-4-enyl]-4,4,10,13,14-pentamethyl-1,2,5,6,9,11,12,15
(5S,9S,10R,13S,14S,17R)-17-[(1R)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.47 |
15.95 |
-4.93 |
0 |
1 |
0 |
17 |
424.713 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5R,9R,10R,13S,14S,17R)-4,4,10,13,14-pentamethyl-17-[5-(2-methylpropanoyl)-1H-pyrrol-3-yl]-1,2,5,6,9
(5R,9R,10R,13S,14S,17R)-4,4,10,1…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.34 |
14.2 |
-12.68 |
1 |
3 |
0 |
50 |
449.679 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5R,9R,10R,13S,14S,17S)-4,4,10,13,14-pentamethyl-17-[(3S,5R)-5-(2-methylprop-1-enyl)tetrahydrofuran-
(5R,9R,10R,13S,14S,17S)-4,4,10,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80482-1-O |
SK-MEL-2 (Melanoma Cells) (cluster #1 Of 4), Other |
Other |
6070 |
0.23 |
Functional ≤ 10μM |
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
3640 |
0.24 |
Functional ≤ 10μM |
|
Z80600-2-O |
XF498 (Glioma Cells) (cluster #2 Of 2), Other |
Other |
5100 |
0.23 |
Functional ≤ 10μM |
|
Z80682-3-O |
A549 (Lung Carcinoma Cells) (cluster #3 Of 11), Other |
Other |
4020 |
0.24 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.54 |
14.29 |
-5.85 |
0 |
2 |
0 |
26 |
438.696 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5S,9R,10R,13S,14S,17S)-4,4,10,13,14-pentamethyl-17-[(3S,5R)-5-(2-methylprop-1-enyl)tetrahydrofuran-
(5S,9R,10R,13S,14S,17S)-4,4,10,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80482-1-O |
SK-MEL-2 (Melanoma Cells) (cluster #1 Of 4), Other |
Other |
6070 |
0.23 |
Functional ≤ 10μM |
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
3640 |
0.24 |
Functional ≤ 10μM |
|
Z80600-2-O |
XF498 (Glioma Cells) (cluster #2 Of 2), Other |
Other |
5100 |
0.23 |
Functional ≤ 10μM |
|
Z80682-3-O |
A549 (Lung Carcinoma Cells) (cluster #3 Of 11), Other |
Other |
4020 |
0.24 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.54 |
14.05 |
-5.94 |
0 |
2 |
0 |
26 |
438.696 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5R,9R,10R,13S,14S,17S)-17-[(E,1S)-5-methoxy-1,5-dimethyl-hex-3-enyl]-4,4,10,13,14-pentamethyl-1,2,5
(5R,9R,10R,13S,14S,17S)-17-[(E,1…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.72 |
15.11 |
-5.4 |
0 |
2 |
0 |
26 |
454.739 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5S,9R,10R,13S,14S,17S)-17-[(E,1S)-5-methoxy-1,5-dimethyl-hex-3-enyl]-4,4,10,13,14-pentamethyl-1,2,5
(5S,9R,10R,13S,14S,17S)-17-[(E,1…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.72 |
14.95 |
-5.39 |
0 |
2 |
0 |
26 |
454.739 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5R,9R,10R,13S,14S,17S)-4,4,10,13,14-pentamethyl-17-[(3S,5S)-5-(2-methylprop-1-enyl)tetrahydrofuran-
(5R,9R,10R,13S,14S,17S)-4,4,10,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80482-1-O |
SK-MEL-2 (Melanoma Cells) (cluster #1 Of 4), Other |
Other |
4730 |
0.23 |
Functional ≤ 10μM |
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
3820 |
0.24 |
Functional ≤ 10μM |
|
Z80600-2-O |
XF498 (Glioma Cells) (cluster #2 Of 2), Other |
Other |
5810 |
0.23 |
Functional ≤ 10μM |
|
Z80682-3-O |
A549 (Lung Carcinoma Cells) (cluster #3 Of 11), Other |
Other |
4290 |
0.23 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.54 |
14.28 |
-6.02 |
0 |
2 |
0 |
26 |
438.696 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5S,9R,10R,13S,14S,17S)-4,4,10,13,14-pentamethyl-17-[(3S,5S)-5-(2-methylprop-1-enyl)tetrahydrofuran-
(5S,9R,10R,13S,14S,17S)-4,4,10,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80482-1-O |
SK-MEL-2 (Melanoma Cells) (cluster #1 Of 4), Other |
Other |
4730 |
0.23 |
Functional ≤ 10μM |
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
3820 |
0.24 |
Functional ≤ 10μM |
|
Z80600-2-O |
XF498 (Glioma Cells) (cluster #2 Of 2), Other |
Other |
5810 |
0.23 |
Functional ≤ 10μM |
|
Z80682-3-O |
A549 (Lung Carcinoma Cells) (cluster #3 Of 11), Other |
Other |
4290 |
0.23 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.54 |
14.08 |
-5.9 |
0 |
2 |
0 |
26 |
438.696 |
2 |
↓
|
|