|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.90 |
7.36 |
-53.6 |
4 |
4 |
1 |
65 |
279.367 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.90 |
7.82 |
-81.32 |
5 |
4 |
2 |
67 |
280.375 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
6.4 |
-8.07 |
3 |
4 |
0 |
64 |
250.305 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.97 |
6.87 |
-28.62 |
4 |
4 |
1 |
65 |
251.313 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
0.73 |
-8.86 |
1 |
3 |
0 |
38 |
303.287 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
-0.96 |
-8.97 |
1 |
4 |
0 |
47 |
265.316 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.22 |
-1.62 |
-15.15 |
2 |
7 |
0 |
87 |
459.472 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
-0.19 |
-19.52 |
2 |
7 |
0 |
93 |
460.456 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
6.82 |
-8.94 |
2 |
4 |
0 |
58 |
279.343 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.48 |
7.19 |
-30.33 |
3 |
4 |
1 |
59 |
280.351 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
6.79 |
-9.17 |
2 |
4 |
0 |
58 |
279.343 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.48 |
7.15 |
-30.59 |
3 |
4 |
1 |
59 |
280.351 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
126 |
0.29 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
175 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
9.85 |
-21.14 |
3 |
8 |
0 |
106 |
456.518 |
12 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.11 |
10.31 |
-47.21 |
4 |
8 |
1 |
107 |
457.526 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
614 |
0.26 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
557 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.62 |
10.3 |
-20.5 |
3 |
8 |
0 |
106 |
472.973 |
12 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.62 |
10.77 |
-46.28 |
4 |
8 |
1 |
107 |
473.981 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
17 |
0.44 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.46 |
Functional ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
231 |
0.37 |
Functional ≤ 10μM
|
|
Z100492-1-O |
MCF10 (Normal Breast Epithelial Cells) (cluster #1 Of 1), Other |
Other |
188 |
0.38 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1433 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.46 |
9.67 |
-9.34 |
1 |
5 |
0 |
56 |
341.386 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
8.29 |
-9.15 |
1 |
5 |
0 |
56 |
329.787 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.22 |
8.76 |
-34.89 |
2 |
5 |
1 |
58 |
330.795 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.82 |
-0.95 |
-9.17 |
1 |
5 |
0 |
56 |
364.232 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.54 |
7.77 |
-8.85 |
1 |
5 |
0 |
56 |
295.342 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.54 |
8.24 |
-32.45 |
2 |
5 |
1 |
58 |
296.35 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
7.06 |
-10.3 |
1 |
6 |
0 |
66 |
325.368 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.57 |
7.53 |
-33.84 |
2 |
6 |
1 |
67 |
326.376 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
48 |
0.45 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
489 |
0.38 |
Functional ≤ 10μM
|
|
Z100492-1-O |
MCF10 (Normal Breast Epithelial Cells) (cluster #1 Of 1), Other |
Other |
489 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.70 |
7.84 |
-9.82 |
1 |
5 |
0 |
56 |
313.332 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.70 |
8.31 |
-35.71 |
2 |
5 |
1 |
58 |
314.34 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.39 |
1.81 |
-10.76 |
1 |
5 |
0 |
56 |
363.339 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
7.8 |
-10.27 |
1 |
5 |
0 |
56 |
313.332 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.66 |
8.27 |
-32.58 |
2 |
5 |
1 |
58 |
314.34 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
8.49 |
-8.7 |
1 |
5 |
0 |
56 |
309.369 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.94 |
8.96 |
-32.51 |
2 |
5 |
1 |
58 |
310.377 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
8.45 |
-8.86 |
1 |
5 |
0 |
56 |
309.369 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.99 |
8.91 |
-32.45 |
2 |
5 |
1 |
58 |
310.377 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
6.01 |
-8.89 |
2 |
4 |
0 |
58 |
299.761 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
6 |
-8.89 |
2 |
4 |
0 |
58 |
299.761 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.61 |
13.31 |
-80.71 |
4 |
7 |
1 |
101 |
366.445 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.61 |
12.83 |
-59.66 |
3 |
7 |
0 |
100 |
365.437 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.61 |
11.2 |
-49.02 |
3 |
7 |
0 |
100 |
365.437 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
6.55 |
-47.63 |
4 |
5 |
1 |
77 |
355.437 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.05 |
6.17 |
-10.92 |
3 |
5 |
0 |
75 |
354.429 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.05 |
7.13 |
-101.92 |
5 |
5 |
2 |
78 |
356.445 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
5.1 |
-47.38 |
4 |
6 |
1 |
84 |
339.419 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.05 |
4.72 |
-10.53 |
3 |
6 |
0 |
82 |
338.411 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.05 |
5.66 |
-99.6 |
5 |
6 |
2 |
85 |
340.427 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.65 |
5.67 |
-49.89 |
4 |
6 |
1 |
84 |
339.419 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.65 |
5.29 |
-11.39 |
3 |
6 |
0 |
82 |
338.411 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.65 |
6.23 |
-105.16 |
5 |
6 |
2 |
85 |
340.427 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
7.63 |
-46.99 |
3 |
5 |
1 |
66 |
323.42 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.31 |
7.25 |
-10.15 |
2 |
5 |
0 |
64 |
322.412 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.31 |
8.19 |
-95.91 |
4 |
5 |
2 |
67 |
324.428 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
6.6 |
-11.51 |
2 |
6 |
0 |
84 |
323.352 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.25 |
8.67 |
-6.64 |
1 |
3 |
0 |
38 |
271.27 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.76 |
9.12 |
-6.05 |
1 |
3 |
0 |
38 |
287.725 |
3 |
↓
|
|
|
|
Analogs
-
41470985
-
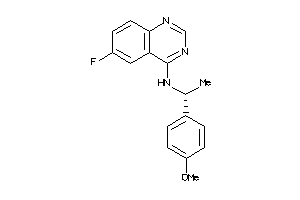
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.70 |
8.76 |
-7.29 |
1 |
4 |
0 |
47 |
297.333 |
4 |
↓
|
|
|
|
Analogs
-
41470983
-
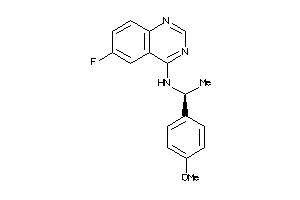
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.70 |
8.74 |
-7.27 |
1 |
4 |
0 |
47 |
297.333 |
4 |
↓
|
|
|
|
Analogs
-
13863972
-
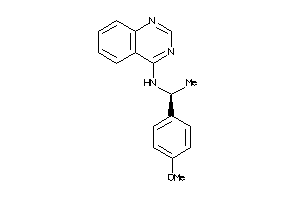
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
8.68 |
-9.14 |
1 |
4 |
0 |
47 |
279.343 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.53 |
9.15 |
-30.8 |
2 |
4 |
1 |
48 |
280.351 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.06 |
9.72 |
-7.65 |
1 |
3 |
0 |
38 |
277.371 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.06 |
10.21 |
-23.28 |
2 |
3 |
1 |
39 |
278.379 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.28 |
10.47 |
-10.48 |
0 |
3 |
0 |
29 |
267.307 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
11.15 |
-7.97 |
0 |
3 |
0 |
29 |
263.344 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.54 |
11.61 |
-29.48 |
1 |
3 |
1 |
30 |
264.352 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
7.81 |
-9.39 |
1 |
4 |
0 |
49 |
279.343 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.53 |
8.28 |
-28.61 |
2 |
4 |
1 |
50 |
280.351 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.35 |
9.56 |
-8.37 |
1 |
3 |
0 |
38 |
281.384 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.35 |
10.02 |
-30.53 |
2 |
3 |
1 |
39 |
282.392 |
4 |
↓
|
|
|
|
Analogs
-
34891247
-
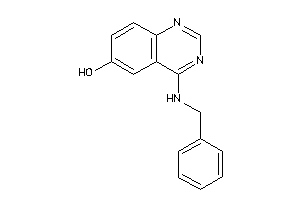
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
5.68 |
-9.52 |
2 |
4 |
0 |
58 |
251.289 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.41 |
6.15 |
-31.01 |
3 |
4 |
1 |
59 |
252.297 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.33 |
9.57 |
-7.66 |
1 |
3 |
0 |
38 |
348.631 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.33 |
10.03 |
-29.91 |
2 |
3 |
1 |
39 |
349.639 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
12.9 |
-45.02 |
0 |
5 |
-1 |
69 |
368.844 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.99 |
13.44 |
-50.25 |
1 |
5 |
0 |
70 |
369.852 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.32 |
8.41 |
-43.42 |
3 |
4 |
1 |
57 |
279.367 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.32 |
8.87 |
-93.66 |
4 |
4 |
2 |
58 |
280.375 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
6.56 |
-53.84 |
4 |
4 |
1 |
65 |
265.34 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.08 |
7.03 |
-86.18 |
5 |
4 |
2 |
67 |
266.348 |
4 |
↓
|
|
|
|
Analogs
-
34568453
-
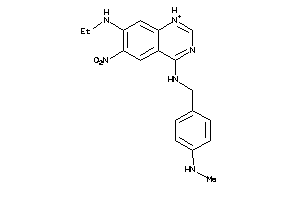
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
9.25 |
-11.88 |
1 |
6 |
0 |
84 |
280.287 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.88 |
9.72 |
-38.47 |
2 |
6 |
1 |
85 |
281.295 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
8.76 |
-8.81 |
1 |
4 |
0 |
47 |
279.343 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.33 |
9.23 |
-29.61 |
2 |
4 |
1 |
48 |
280.351 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.99 |
6.41 |
-8.23 |
3 |
4 |
0 |
64 |
250.305 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.99 |
6.88 |
-28.36 |
4 |
4 |
1 |
65 |
251.313 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.91 |
3.58 |
-10.09 |
4 |
5 |
0 |
84 |
266.304 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.91 |
4.04 |
-30.25 |
5 |
5 |
1 |
85 |
267.312 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.59 |
9.02 |
-8.47 |
2 |
4 |
0 |
55 |
278.359 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.59 |
9.48 |
-29.99 |
3 |
4 |
1 |
56 |
279.367 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.61 |
9.03 |
-8.69 |
2 |
4 |
0 |
55 |
278.359 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.61 |
9.49 |
-29.55 |
3 |
4 |
1 |
56 |
279.367 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
8.99 |
-6.72 |
2 |
4 |
0 |
55 |
278.359 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.97 |
9.46 |
-30.31 |
3 |
4 |
1 |
56 |
279.367 |
4 |
↓
|
|