|
|
Analogs
-
40918437
-
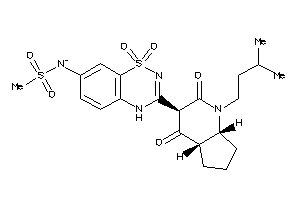
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.69 |
3.38 |
-29.83 |
1 |
10 |
0 |
144 |
495.603 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.28 |
2.74 |
-217.75 |
0 |
10 |
-3 |
152 |
493.587 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.69 |
2.98 |
-25.55 |
0 |
10 |
0 |
146 |
494.595 |
6 |
↓
|
|
|
|
Analogs
-
40918437
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 5), Other |
Other |
9 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
3.16 |
-27.1 |
1 |
10 |
0 |
144 |
509.63 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.60 |
3.12 |
-218.33 |
0 |
10 |
-3 |
152 |
507.614 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.01 |
2.84 |
-23.25 |
0 |
10 |
0 |
146 |
508.622 |
6 |
↓
|
|
|
|
Analogs
-
40918437
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 5), Other |
Other |
24 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
3.7 |
-23.53 |
1 |
10 |
0 |
144 |
509.63 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.60 |
3.06 |
-214.32 |
0 |
10 |
-3 |
152 |
507.614 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.01 |
3.26 |
-20.6 |
0 |
10 |
0 |
146 |
508.622 |
6 |
↓
|
|
|
|
Analogs
-
40916675
-
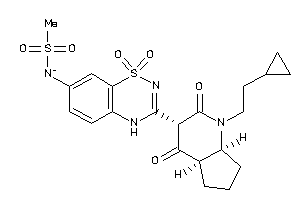
-
40916677
-
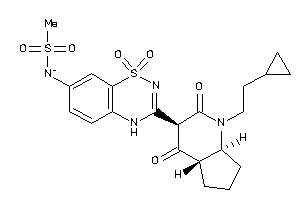
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 5), Other |
Other |
24 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
3.92 |
-28.6 |
1 |
10 |
0 |
144 |
509.63 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.60 |
3.15 |
-216.36 |
0 |
10 |
-3 |
152 |
507.614 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.01 |
3.3 |
-27.88 |
1 |
10 |
0 |
144 |
509.63 |
6 |
↓
|
|
|
|
Analogs
-
40916675
-

-
40916677
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 5), Other |
Other |
24 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
3.18 |
-27.43 |
1 |
10 |
0 |
144 |
509.63 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.60 |
3.35 |
-218.17 |
0 |
10 |
-3 |
152 |
507.614 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.01 |
2.81 |
-24.56 |
0 |
10 |
0 |
146 |
508.622 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 5), Other |
Other |
2100 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.42 |
1.34 |
-29.76 |
1 |
10 |
0 |
144 |
453.522 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.00 |
0.68 |
-215.23 |
0 |
10 |
-3 |
152 |
451.506 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.42 |
0.94 |
-25.58 |
0 |
10 |
0 |
146 |
452.514 |
4 |
↓
|
|
|
|
Analogs
-
40915798
-
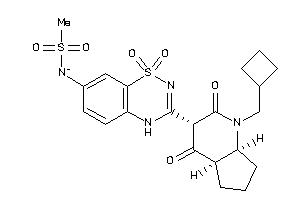
-
40915800
-
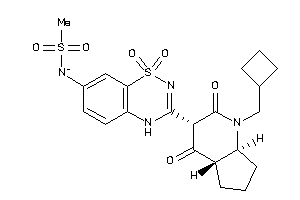
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 5), Other |
Other |
2100 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.42 |
1.49 |
-28.57 |
1 |
10 |
0 |
144 |
453.522 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.00 |
0.72 |
-213.14 |
0 |
10 |
-3 |
152 |
451.506 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.42 |
1.04 |
-24.42 |
0 |
10 |
0 |
146 |
452.514 |
4 |
↓
|
|
|
|
Analogs
-
40915798
-

-
40915800
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 5), Other |
Other |
2100 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.42 |
1.38 |
-30.27 |
1 |
10 |
0 |
144 |
453.522 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.00 |
0.91 |
-215.28 |
0 |
10 |
-3 |
152 |
451.506 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.42 |
0.98 |
-24.38 |
0 |
10 |
0 |
146 |
452.514 |
4 |
↓
|
|
|
|
Analogs
-
40916675
-

-
40916677
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 5), Other |
Other |
60 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.69 |
3.4 |
-23.9 |
1 |
10 |
0 |
144 |
495.603 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.28 |
2.76 |
-215.24 |
0 |
10 |
-3 |
152 |
493.587 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.69 |
3 |
-19.92 |
0 |
10 |
0 |
146 |
494.595 |
6 |
↓
|
|
|
|
Analogs
-
40916675
-

-
40916677
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 5), Other |
Other |
60 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.69 |
3.43 |
-30.46 |
1 |
10 |
0 |
144 |
495.603 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.28 |
2.97 |
-217.48 |
0 |
10 |
-3 |
152 |
493.587 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.69 |
3.03 |
-24.27 |
0 |
10 |
0 |
146 |
494.595 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.71 |
1.92 |
-23.05 |
1 |
10 |
0 |
144 |
467.549 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.30 |
1.27 |
-212.96 |
0 |
10 |
-3 |
152 |
465.533 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.71 |
1.48 |
-20.1 |
0 |
10 |
0 |
146 |
466.541 |
4 |
↓
|
|
|
|
Analogs
-
53122849
-
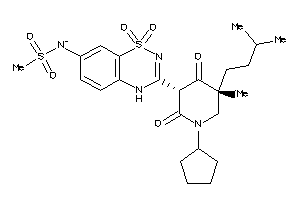
-
53122852
-
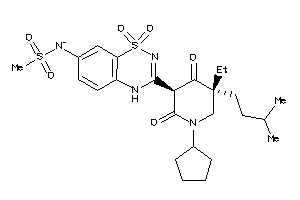
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.71 |
1.92 |
-29.61 |
1 |
10 |
0 |
144 |
467.549 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.30 |
1.34 |
-215.03 |
0 |
10 |
-3 |
152 |
465.533 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.71 |
1.52 |
-23.51 |
0 |
10 |
0 |
146 |
466.541 |
4 |
↓
|
|