|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4000 |
0.24 |
Binding ≤ 10μM
|
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.34 |
Binding ≤ 10μM
|
|
FDFT-1-E |
Squalene Synthetase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.23 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.24 |
Binding ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3200 |
0.25 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
THA-1-E |
Thyroid Hormone Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
650 |
0.28 |
Binding ≤ 10μM
|
|
THB-1-E |
Thyroid Hormone Receptor Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.28 |
Binding ≤ 10μM
|
|
CP2C8-1-E |
Cytochrome P450 2C8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.26 |
ADME/T ≤ 10μM
|
|
LEF-2-B |
Anthrax Lethal Factor (cluster #2 Of 2), Bacterial |
Bacteria |
3500 |
0.25 |
Binding ≤ 10μM
|
|
ERG2-2-F |
C-8 Sterol Isomerase (cluster #2 Of 2), Fungal |
Fungi |
62 |
0.33 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
2500 |
0.25 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
9000 |
0.23 |
Functional ≤ 10μM
|
|
Z80773-1-O |
CHRC/5 Cell Line (cluster #1 Of 1), Other |
Other |
2300 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.36 |
2.42 |
-36.05 |
1 |
4 |
1 |
43 |
646.327 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.25 |
Binding ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.25 |
Binding ≤ 10μM
|
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.16 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.92 |
17.7 |
-43.27 |
1 |
10 |
0 |
124 |
610.711 |
11 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.67 |
18.37 |
-45.36 |
2 |
10 |
1 |
118 |
611.719 |
12 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.67 |
16.67 |
-18.95 |
1 |
10 |
0 |
117 |
610.711 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.25 |
Binding ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.25 |
Binding ≤ 10μM
|
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.16 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.92 |
18.05 |
-40.73 |
1 |
10 |
0 |
124 |
610.711 |
11 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.67 |
18.5 |
-52.04 |
2 |
10 |
1 |
118 |
611.719 |
12 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.67 |
16.87 |
-16.27 |
1 |
10 |
0 |
117 |
610.711 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-2-E |
Cruzipain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.35 |
Binding ≤ 10μM
|
|
Q95PM0-2-E |
Rhodesain (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8000 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
7.64 |
-17.27 |
3 |
4 |
0 |
62 |
285.372 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
3.55 |
7.85 |
-12.77 |
3 |
4 |
0 |
60 |
285.372 |
5 |
↓
|
|
|
|
Analogs
-
9580294
-
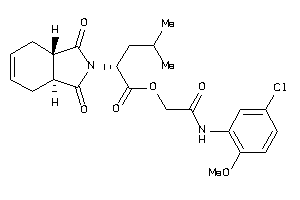
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
10.69 |
-13.47 |
1 |
8 |
0 |
102 |
462.93 |
9 |
↓
|
|
|
|
Analogs
-
8691187
-
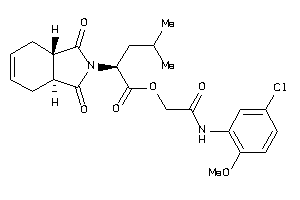
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
10.77 |
-14.74 |
1 |
8 |
0 |
102 |
462.93 |
9 |
↓
|
|
|
|
Analogs
-
3363859
-
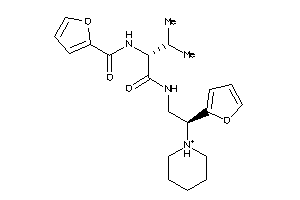
-
3363862
-
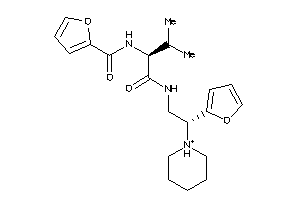
-
3363866
-
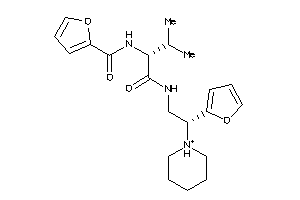
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7000 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
7.29 |
-44.6 |
3 |
7 |
1 |
89 |
388.488 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.57 |
5.37 |
-24.47 |
2 |
7 |
0 |
95 |
387.48 |
8 |
↓
|
|
|
|
Analogs
-
3363862
-

-
3363866
-

-
3363856
-
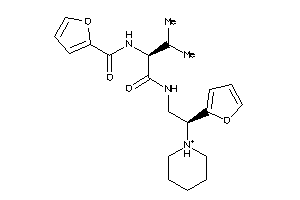
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7000 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
7.64 |
-43.04 |
3 |
7 |
1 |
89 |
388.488 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.57 |
5.2 |
-35.8 |
2 |
7 |
0 |
95 |
387.48 |
8 |
↓
|
|
|
|
Analogs
-
3363866
-

-
3363856
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7000 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
7.69 |
-41.46 |
3 |
7 |
1 |
89 |
388.488 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.57 |
5.31 |
-29.3 |
2 |
7 |
0 |
95 |
387.48 |
8 |
↓
|
|
|
|
Analogs
-
3363856
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7000 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
7.66 |
-42.68 |
3 |
7 |
1 |
89 |
388.488 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.57 |
5.67 |
-33.74 |
2 |
7 |
0 |
95 |
387.48 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
700 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.69 |
11.87 |
-16.29 |
1 |
6 |
0 |
82 |
391.423 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
8.01 |
-14.62 |
1 |
7 |
0 |
91 |
389.791 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
11.89 |
-13.47 |
2 |
6 |
0 |
85 |
490.865 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
11.89 |
-13.46 |
2 |
6 |
0 |
85 |
490.865 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
0.83 |
-16.29 |
3 |
9 |
0 |
126 |
464.453 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
9.53 |
-16.11 |
3 |
9 |
0 |
126 |
464.453 |
9 |
↓
|
|
|
|
Analogs
-
8693977
-
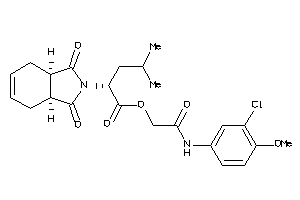
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
800 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
10.66 |
-17.6 |
1 |
8 |
0 |
102 |
462.93 |
9 |
↓
|
|
|
|
Analogs
-
8693976
-
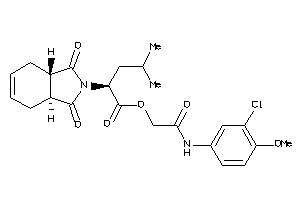
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
800 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
11.26 |
-16.34 |
1 |
8 |
0 |
102 |
462.93 |
9 |
↓
|
|
|
|
Analogs
-
12819559
-
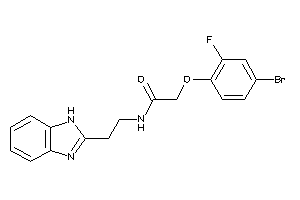
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-3-E |
Cruzipain (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
7.31 |
-22.87 |
2 |
5 |
0 |
67 |
374.238 |
6 |
↓
|
|
|
|
Analogs
-
5093465
-
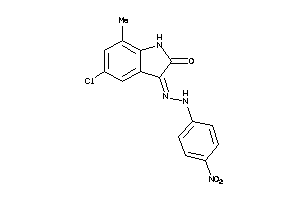
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-3-E |
Cruzipain (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.45 |
Binding ≤ 10μM
|
|
Q9N6S8-1-E |
Falcipain 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9400 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
3.3 |
-12.39 |
4 |
5 |
0 |
83 |
268.729 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.60 |
1.48 |
-40.21 |
3 |
5 |
-1 |
86 |
267.721 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.23 |
1.66 |
-32.4 |
4 |
5 |
0 |
86 |
268.729 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-3-E |
Cruzipain (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
1.54 |
-39.32 |
5 |
5 |
1 |
88 |
347.161 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.09 |
2.36 |
-13.98 |
4 |
5 |
0 |
91 |
346.153 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.63 |
1.41 |
-39.54 |
3 |
5 |
-1 |
86 |
345.145 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-3-E |
Cruzipain (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.43 |
14.23 |
-23.36 |
2 |
4 |
1 |
44 |
474.415 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.32 |
9.18 |
-41.96 |
3 |
9 |
1 |
125 |
482.52 |
8 |
↓
|
|
Ref
Reference (pH 7)
|
5.32 |
9.03 |
-36.32 |
3 |
9 |
1 |
125 |
482.52 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.32 |
9.8 |
-16.61 |
2 |
9 |
0 |
127 |
481.512 |
8 |
↓
|
|
|
|
Analogs
-
4097344
-
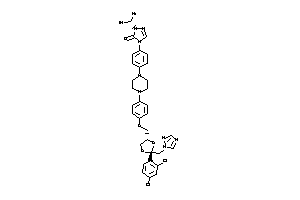
-
3830973
-
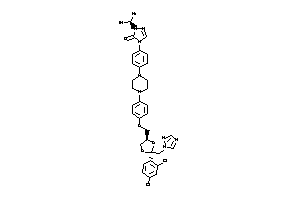
-
3830974
-
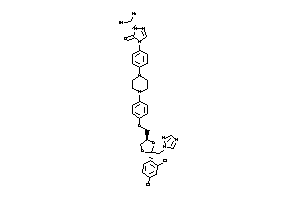
-
3830975
-
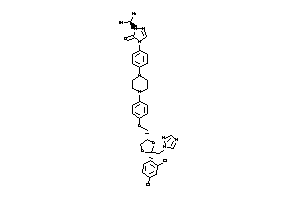
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP51A-1-E |
Cytochrome P450 51 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.16 |
Binding ≤ 10μM
|
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.16 |
Binding ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.21 |
Functional ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.19 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
700 |
0.18 |
ADME/T ≤ 10μM
|
|
Q96W81-1-F |
14-alpha Sterol Demethylase (cluster #1 Of 1), Fungal |
Fungi |
31 |
0.21 |
Binding ≤ 10μM
|
|
Q4WNT5-1-O |
14-alpha Sterol Demethylase Cyp51A (cluster #1 Of 1), Other |
Other |
1010 |
0.17 |
Binding ≤ 10μM
|
|
Z102121-2-O |
Trichophyton Mentagrophytes (cluster #2 Of 3), Other |
Other |
370 |
0.18 |
Functional ≤ 10μM
|
|
Z50046-1-O |
Trichophyton Quinckeanum (cluster #1 Of 2), Other |
Other |
2930 |
0.16 |
Functional ≤ 10μM
|
|
Z50408-1-O |
Issatchenkia Orientalis (cluster #1 Of 2), Other |
Other |
4210 |
0.15 |
Functional ≤ 10μM
|
|
Z50409-1-O |
Kluyveromyces Marxianus (cluster #1 Of 2), Other |
Other |
400 |
0.18 |
Functional ≤ 10μM
|
|
Z50442-1-O |
Candida Albicans (cluster #1 Of 4), Other |
Other |
1410 |
0.17 |
Functional ≤ 10μM
|
|
Z50443-1-O |
Candida Glabrata (cluster #1 Of 1), Other |
Other |
3740 |
0.16 |
Functional ≤ 10μM
|
|
Z50452-1-O |
Trichophyton Rubrum (cluster #1 Of 2), Other |
Other |
980 |
0.17 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.32 |
-1.71 |
-21.17 |
0 |
12 |
0 |
104 |
705.647 |
11 |
↓
|
|
|
|
Analogs
-
4097343
-
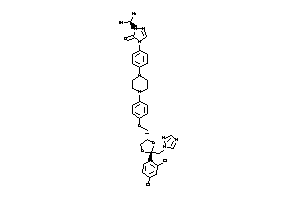
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP51A-1-E |
Cytochrome P450 51 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.16 |
Binding ≤ 10μM
|
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.16 |
Binding ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.21 |
Functional ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.19 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
700 |
0.18 |
ADME/T ≤ 10μM
|
|
Q96W81-1-F |
14-alpha Sterol Demethylase (cluster #1 Of 1), Fungal |
Fungi |
31 |
0.21 |
Binding ≤ 10μM
|
|
Q4WNT5-1-O |
14-alpha Sterol Demethylase Cyp51A (cluster #1 Of 1), Other |
Other |
1010 |
0.17 |
Binding ≤ 10μM
|
|
Z102121-2-O |
Trichophyton Mentagrophytes (cluster #2 Of 3), Other |
Other |
370 |
0.18 |
Functional ≤ 10μM
|
|
Z50046-1-O |
Trichophyton Quinckeanum (cluster #1 Of 2), Other |
Other |
2930 |
0.16 |
Functional ≤ 10μM
|
|
Z50408-1-O |
Issatchenkia Orientalis (cluster #1 Of 2), Other |
Other |
4210 |
0.15 |
Functional ≤ 10μM
|
|
Z50409-1-O |
Kluyveromyces Marxianus (cluster #1 Of 2), Other |
Other |
400 |
0.18 |
Functional ≤ 10μM
|
|
Z50442-1-O |
Candida Albicans (cluster #1 Of 4), Other |
Other |
1410 |
0.17 |
Functional ≤ 10μM
|
|
Z50443-1-O |
Candida Glabrata (cluster #1 Of 1), Other |
Other |
3740 |
0.16 |
Functional ≤ 10μM
|
|
Z50452-1-O |
Trichophyton Rubrum (cluster #1 Of 2), Other |
Other |
980 |
0.17 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.32 |
-1.71 |
-21.17 |
0 |
12 |
0 |
104 |
705.647 |
11 |
↓
|
|
|
|
Analogs
-
378484
-
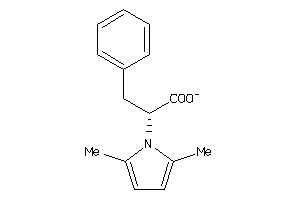
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.40 |
9.85 |
-45.87 |
0 |
3 |
-1 |
45 |
242.298 |
4 |
↓
|
|
|
|
Analogs
-
378481
-
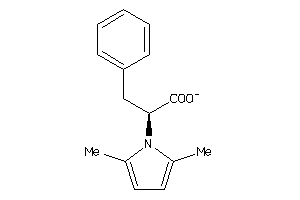
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.40 |
2.09 |
-44.97 |
0 |
3 |
-1 |
45 |
242.298 |
4 |
↓
|
|
|
|
Analogs
-
13758259
-
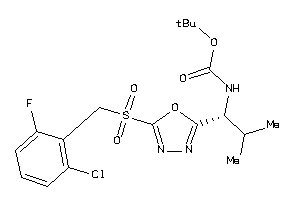
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.28 |
1.63 |
-11.97 |
1 |
8 |
0 |
111 |
447.916 |
8 |
↓
|
|
|
|
Analogs
-
4952976
-
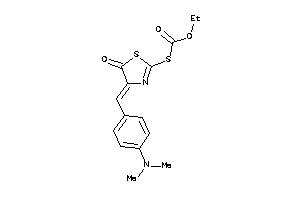
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.08 |
8.01 |
-9.22 |
0 |
5 |
0 |
60 |
336.438 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABCG2-2-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.26 |
Binding ≤ 10μM
|
|
CYSP-4-E |
Cruzipain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
4000 |
0.27 |
Binding ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7600 |
0.26 |
Binding ≤ 10μM
|
|
MRP1-1-E |
Multidrug Resistance-associated Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8500 |
0.25 |
Binding ≤ 10μM
|
|
MRP2-1-E |
Canalicular Multispecific Organic Anion Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
MRP3-1-E |
Canalicular Multispecific Organic Anion Transporter 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5100 |
0.26 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
726 |
0.31 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
10 |
0.40 |
Functional ≤ 10μM
|
|
Z50677-1-O |
Human Immunodeficiency Virus (cluster #1 Of 3), Other |
Other |
9 |
0.40 |
Functional ≤ 10μM
|
|
Z80295-6-O |
MT4 (Lymphocytes) (cluster #6 Of 8), Other |
Other |
7 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.03 |
0.49 |
-10.21 |
3 |
7 |
0 |
120 |
435.285 |
4 |
↓
|
|
|
|
Analogs
-
4401071
-
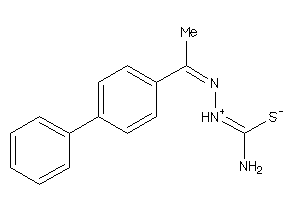
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
8.41 |
-12.09 |
3 |
3 |
0 |
50 |
269.373 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.95 |
7.85 |
-14.96 |
0 |
4 |
0 |
48 |
281.311 |
3 |
↓
|
|
|
|
Analogs
-
33309810
-
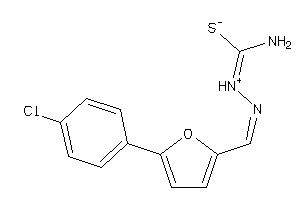
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-2-E |
Cruzipain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
6.71 |
-13.65 |
3 |
4 |
0 |
64 |
279.752 |
4 |
↓
|
|
|
|
Analogs
-
33306174
-
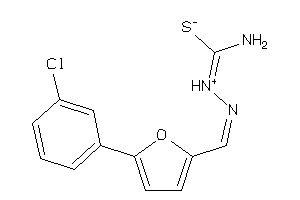
-
35495957
-
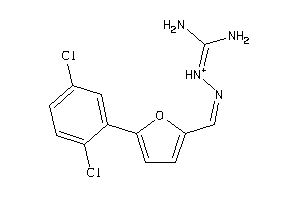
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-2-E |
Cruzipain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.71 |
6.7 |
-13.12 |
3 |
4 |
0 |
64 |
279.752 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-2-E |
Cruzipain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
170 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
6 |
-12.95 |
3 |
3 |
0 |
50 |
261.272 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
39977976
-
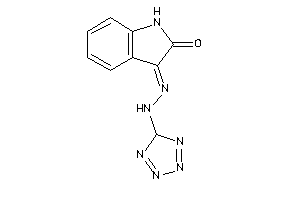
-
13122245
-
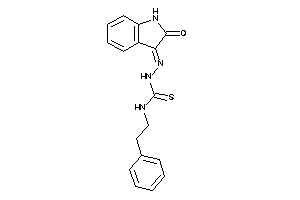
Draw
Identity
99%
90%
80%
70%
Vendors
And 43 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-3-E |
Cruzipain (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
2.14 |
-14.08 |
4 |
5 |
0 |
83 |
220.257 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
0.94 |
2.18 |
-36.83 |
5 |
5 |
1 |
81 |
221.265 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
1.57 |
-0.72 |
-10.98 |
4 |
5 |
0 |
84 |
220.257 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.57 |
13.78 |
-24.53 |
2 |
7 |
0 |
104 |
499.592 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.57 |
13.75 |
-20.19 |
2 |
7 |
0 |
104 |
499.592 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-2-E |
Cruzipain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
460 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.17 |
5.42 |
-13.06 |
3 |
3 |
0 |
50 |
248.138 |
3 |
↓
|
|
|
|
Analogs
-
38582598
-
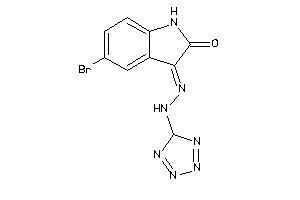
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-3-E |
Cruzipain (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7000 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.81 |
1.06 |
-39.21 |
5 |
5 |
1 |
88 |
300.161 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
1.90 |
2.62 |
-12.39 |
4 |
5 |
0 |
83 |
299.153 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.81 |
1.87 |
-14.21 |
4 |
5 |
0 |
91 |
299.153 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-2-E |
Cruzipain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
560 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
5.26 |
-14.95 |
3 |
3 |
0 |
52 |
248.138 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-4-E |
Cruzipain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
710 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
9.22 |
-5.66 |
2 |
6 |
0 |
87 |
240.27 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.30 |
9.43 |
-10.66 |
1 |
6 |
0 |
79 |
300.272 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-4-E |
Cruzipain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
90 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
5.82 |
-4.62 |
0 |
6 |
0 |
75 |
233.275 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-4-E |
Cruzipain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
12.01 |
-7.24 |
2 |
6 |
0 |
92 |
288.314 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-3-E |
Cruzipain (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.64 |
5.21 |
-10.36 |
2 |
6 |
0 |
66 |
345.834 |
5 |
↓
|
|
|
|
Analogs
-
37858367
-
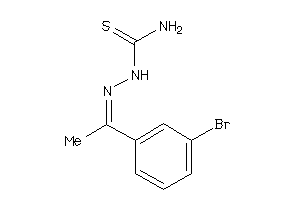
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-2-E |
Cruzipain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
60 |
0.72 |
Binding ≤ 10μM
|
|
Q95PM0-2-E |
Rhodesain (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.73 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
5.4 |
-17.45 |
3 |
3 |
0 |
52 |
272.171 |
2 |
↓
|
|
|
|
Analogs
-
374541
-
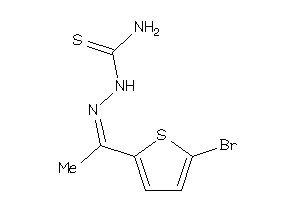
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-2-E |
Cruzipain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3800 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
4.63 |
-13.37 |
3 |
3 |
0 |
52 |
278.2 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYSP-2-E |
Cruzipain (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
280 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.38 |
6.6 |
-20.33 |
3 |
4 |
0 |
65 |
324.203 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
3.87 |
6.82 |
-13.57 |
3 |
4 |
0 |
64 |
324.203 |
4 |
↓
|
|