|
|
Analogs
-
11616941
-
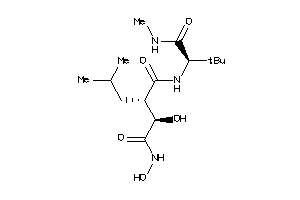
-
11616943
-
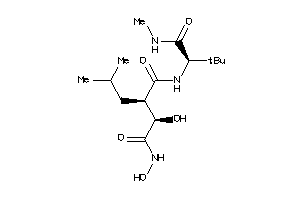
-
27079413
-
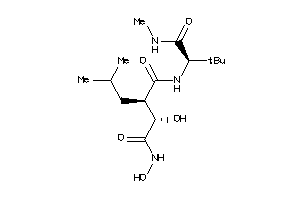
-
43771549
-
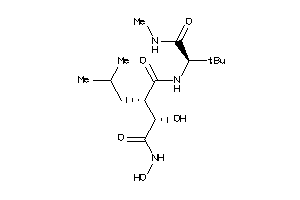
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA17-1-E |
ADAM17 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM
|
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM
|
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
MMP14-3-E |
Matrix Metalloproteinase 14 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
84 |
0.43 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
200 |
0.41 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.47 |
Binding ≤ 10μM
|
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.49 |
Binding ≤ 10μM
|
|
ADA17-1-E |
ADAM17 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.31 |
Functional ≤ 10μM
|
|
TNFA-2-E |
TNF-alpha (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1001 |
0.37 |
Functional ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
4100 |
0.33 |
Functional ≤ 10μM
|
|
Z80548-2-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #2 Of 5), Other |
Other |
2100 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.38 |
0.45 |
-15.72 |
5 |
8 |
0 |
128 |
331.413 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.38 |
1.66 |
-56.16 |
4 |
8 |
-1 |
131 |
330.405 |
8 |
↓
|
|
|
|
Analogs
-
1544157
-
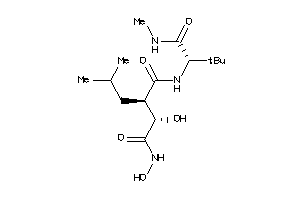
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
200 |
0.41 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.47 |
Binding ≤ 10μM
|
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.38 |
0.65 |
-22.37 |
5 |
8 |
0 |
128 |
331.413 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.38 |
1.88 |
-61.38 |
4 |
8 |
-1 |
131 |
330.405 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
24 |
0.31 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.32 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.31 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
4.04 |
-23.96 |
2 |
11 |
0 |
143 |
513.594 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.09 |
5.37 |
-87.93 |
1 |
11 |
-1 |
145 |
512.586 |
7 |
↓
|
|
|
|
Analogs
-
3979014
-
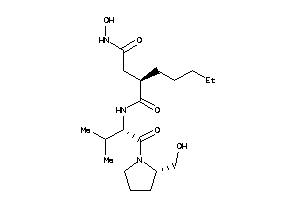
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.34 |
Binding ≤ 10μM |
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM |
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
190 |
0.35 |
Binding ≤ 10μM |
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
330 |
0.34 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
2.49 |
-16.23 |
4 |
8 |
0 |
119 |
385.505 |
11 |
↓
|
|
|
|
Analogs
-
3871393
-
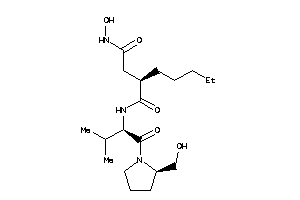
-
3871392
-
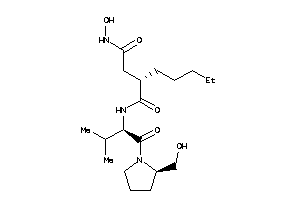
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DEF-1-B |
Peptide Deformylase (cluster #1 Of 1), Bacterial |
Bacteria |
5 |
0.43 |
Binding ≤ 10μM
|
|
Q9JN24-1-B |
Peptide Deformylase (cluster #1 Of 1), Bacterial |
Bacteria |
140 |
0.36 |
Binding ≤ 10μM |
|
DEF1A-1-E |
Peptide Deformylase 1A, Chloroplastic (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.39 |
Binding ≤ 10μM
|
|
DEFM-1-E |
Peptide Deformylase Mitochondrial (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.32 |
Binding ≤ 10μM
|
|
ECE1-1-E |
Endothelin-converting Enzyme 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.28 |
Binding ≤ 10μM
|
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.34 |
Binding ≤ 10μM |
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1100 |
0.31 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.29 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM |
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.27 |
Binding ≤ 10μM
|
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
190 |
0.35 |
Binding ≤ 10μM |
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
330 |
0.34 |
Binding ≤ 10μM |
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8200 |
0.26 |
Binding ≤ 10μM
|
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
2512 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.51 |
2.32 |
-16.44 |
4 |
8 |
0 |
119 |
385.505 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.51 |
3.4 |
-55.95 |
3 |
8 |
-1 |
122 |
384.497 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
43 |
0.29 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
23 |
0.31 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
931 |
0.24 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
6.1 |
-19.67 |
2 |
9 |
0 |
116 |
495.557 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.52 |
5.08 |
-54.23 |
1 |
9 |
-1 |
123 |
494.549 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.33 |
7.31 |
-61.73 |
1 |
9 |
-1 |
119 |
494.549 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
43 |
0.29 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
23 |
0.31 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
931 |
0.24 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
5.94 |
-24.17 |
2 |
9 |
0 |
116 |
495.557 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.52 |
4.49 |
-54.66 |
1 |
9 |
-1 |
123 |
494.549 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.33 |
7.16 |
-63.52 |
1 |
9 |
-1 |
119 |
494.549 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
46 |
0.47 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3700 |
0.35 |
Binding ≤ 10μM
|
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
250 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
2.85 |
-8.43 |
3 |
5 |
0 |
78 |
332.51 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
490 |
0.32 |
Binding ≤ 10μM
|
|
MMP12-1-E |
Macrophage Metalloelastase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MMP13-3-E |
Matrix Metalloproteinase 13 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Binding ≤ 10μM
|
|
MMP14-2-E |
Matrix Metalloproteinase 14 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
MMP16-1-E |
Matrix Metalloproteinase 16 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
51 |
0.36 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.43 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.37 |
Binding ≤ 10μM
|
|
MMP8-4-E |
Matrix Metalloproteinase 8 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.43 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
20 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.64 |
7.24 |
-15.89 |
2 |
7 |
0 |
96 |
406.504 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.83 |
5.88 |
-59.91 |
1 |
7 |
-1 |
102 |
405.496 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.64 |
8.44 |
-65.01 |
1 |
7 |
-1 |
99 |
405.496 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2900 |
0.31 |
Binding ≤ 10μM
|
|
MMP13-3-E |
Matrix Metalloproteinase 13 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
45 |
0.41 |
Binding ≤ 10μM
|
|
MMP14-2-E |
Matrix Metalloproteinase 14 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2300 |
0.32 |
Binding ≤ 10μM
|
|
MMP16-1-E |
Matrix Metalloproteinase 16 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
900 |
0.34 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
13 |
0.44 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5900 |
0.29 |
Binding ≤ 10μM
|
|
MMP8-3-E |
Matrix Metalloproteinase 8 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
260 |
0.37 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
200 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
4.59 |
-15.92 |
2 |
7 |
0 |
96 |
364.423 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.53 |
5.81 |
-53.13 |
1 |
7 |
-1 |
99 |
363.415 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA10-1-E |
ADAM10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
ADA12-1-E |
ADAM12 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Binding ≤ 10μM
|
|
ADA17-1-E |
ADAM17 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
ADAM9-1-E |
ADAM9 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
56 |
0.36 |
Binding ≤ 10μM
|
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Binding ≤ 10μM
|
|
MMP10-1-E |
Matrix Metalloproteinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.38 |
Binding ≤ 10μM
|
|
MMP11-1-E |
Matrix Metalloproteinase 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.38 |
Binding ≤ 10μM
|
|
MMP12-1-E |
Macrophage Metalloelastase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
MMP14-1-E |
Matrix Metalloproteinase 14 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
MMP15-1-E |
Matrix Metalloproteinase 15 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Binding ≤ 10μM
|
|
MMP16-1-E |
Matrix Metalloproteinase 16 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
MMP17-1-E |
Matrix Metalloproteinase 17 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.43 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
|
MMP26-1-E |
Matrix Metalloproteinase 26 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.39 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
33 |
0.37 |
Binding ≤ 10μM
|
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.55 |
3.16 |
-20.09 |
5 |
8 |
0 |
123 |
388.468 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.55 |
4.38 |
-63.44 |
4 |
8 |
-1 |
126 |
387.46 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA17-1-E |
ADAM17 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.31 |
Binding ≤ 10μM
|
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
21 |
0.28 |
Binding ≤ 10μM
|
|
MMP10-1-E |
Matrix Metalloproteinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
190 |
0.25 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.29 |
Binding ≤ 10μM
|
|
MMP14-1-E |
Matrix Metalloproteinase 14 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.22 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
660 |
0.23 |
Binding ≤ 10μM
|
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.31 |
Binding ≤ 10μM
|
|
A4-1-E |
Beta Amyloid A4 Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.25 |
Functional ≤ 10μM
|
|
Z80088-1-O |
CHO (Ovarian Cells) (cluster #1 Of 4), Other |
Other |
200 |
0.25 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
500 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.41 |
12.04 |
-28.44 |
3 |
8 |
0 |
108 |
523.674 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.24 |
1.79 |
-22.89 |
3 |
8 |
0 |
116 |
414.505 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.24 |
3 |
-63.97 |
2 |
8 |
-1 |
119 |
413.497 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.43 |
1.42 |
-55.55 |
2 |
8 |
-1 |
122 |
413.497 |
5 |
↓
|
|
|
|
Analogs
-
6116142
-
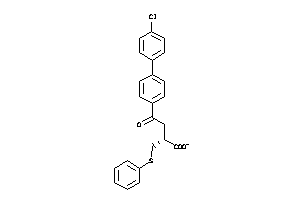
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
MMP13-4-E |
Matrix Metalloproteinase 13 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1470 |
0.29 |
Binding ≤ 10μM
|
|
MMP13-4-E |
Matrix Metalloproteinase 13 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1570 |
0.29 |
Binding ≤ 10μM
|
|
MMP2-2-E |
72 KDa Type IV Collagenase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.40 |
Binding ≤ 10μM
|
|
MMP2-2-E |
72 KDa Type IV Collagenase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
91 |
0.35 |
Binding ≤ 10μM
|
|
MMP3-2-E |
Matrix Metalloproteinase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
143 |
0.34 |
Binding ≤ 10μM
|
|
MMP3-2-E |
Matrix Metalloproteinase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7950 |
0.25 |
Binding ≤ 10μM
|
|
MMP8-4-E |
Matrix Metalloproteinase 8 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
303 |
0.33 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
301 |
0.33 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3110 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.88 |
1.92 |
-59.71 |
0 |
3 |
-1 |
57 |
409.914 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LEF-1-B |
Anthrax Lethal Factor (cluster #1 Of 2), Bacterial |
Bacteria |
1200 |
0.31 |
Binding ≤ 10μM
|
|
ADA10-1-E |
ADAM10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
890 |
0.31 |
Binding ≤ 10μM
|
|
ADA17-2-E |
ADAM17 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
|
BMP1-1-E |
Bone Morphogenetic Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.29 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
8600 |
0.26 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
96 |
0.36 |
Binding ≤ 10μM
|
|
MMP12-1-E |
Macrophage Metalloelastase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
MMP13-3-E |
Matrix Metalloproteinase 13 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
MMP14-2-E |
Matrix Metalloproteinase 14 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.42 |
Binding ≤ 10μM
|
|
MMP16-1-E |
Matrix Metalloproteinase 16 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.42 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
15 |
0.41 |
Binding ≤ 10μM
|
|
MMP25-1-E |
Matrix Metalloproteinase-25 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.45 |
Binding ≤ 10μM |
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.42 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
736 |
0.32 |
Binding ≤ 10μM
|
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM |
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.41 |
Binding ≤ 10μM
|
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 2), Other |
Other |
50 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
3.65 |
-24.43 |
2 |
8 |
0 |
109 |
393.465 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.55 |
1.28 |
-51.2 |
1 |
8 |
-1 |
115 |
392.457 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.37 |
3.89 |
-61.96 |
1 |
8 |
-1 |
112 |
392.457 |
8 |
↓
|
|
|
|
Analogs
-
11616934
-
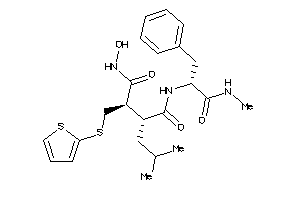
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA17-1-E |
ADAM17 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.34 |
Binding ≤ 10μM
|
|
FCER2-1-E |
Immunoglobulin Epsilon Fc Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.31 |
Binding ≤ 10μM
|
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Binding ≤ 10μM
|
|
MMP14-1-E |
Matrix Metalloproteinase 14 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
47 |
0.32 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.37 |
Binding ≤ 10μM |
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
20 |
0.34 |
Binding ≤ 10μM |
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.36 |
Binding ≤ 10μM |
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.50 |
-4.65 |
-17.84 |
4 |
7 |
0 |
107 |
477.652 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.69 |
-4.09 |
-61.6 |
3 |
7 |
-1 |
113 |
476.644 |
12 |
↓
|
|
|
|
Analogs
-
26639816
-
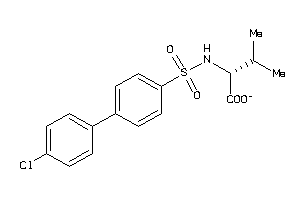
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6500 |
0.30 |
Binding ≤ 10μM
|
|
MMP13-2-E |
Matrix Metalloproteinase 13 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
48 |
0.43 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.46 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.33 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.47 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7500 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
7.54 |
-42.93 |
1 |
5 |
-1 |
86 |
366.846 |
6 |
↓
|
|
|
|
Analogs
-
600699
-
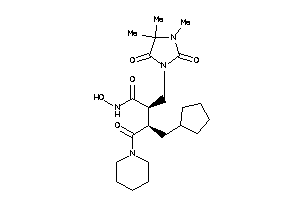
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
527 |
0.28 |
Binding ≤ 10μM |
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 2), Other |
Other |
100 |
0.32 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
-1.05 |
-17.22 |
2 |
9 |
0 |
110 |
436.553 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.07 |
-0.49 |
-66.15 |
1 |
9 |
-1 |
113 |
435.545 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.22 |
1.23 |
-22.17 |
2 |
7 |
0 |
96 |
316.379 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.22 |
2.41 |
-64.1 |
1 |
7 |
-1 |
99 |
315.371 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.74 |
Binding ≤ 10μM
|
|
MMP2-3-E |
72 KDa Type IV Collagenase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
80 |
0.83 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.10 |
-2.45 |
-127.32 |
1 |
5 |
-2 |
92 |
191.123 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
59 |
0.36 |
Binding ≤ 10μM
|
|
MMP13-3-E |
Matrix Metalloproteinase 13 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
2.33 |
-18.46 |
3 |
8 |
0 |
114 |
426.878 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.86 |
0.92 |
-41.61 |
2 |
8 |
-1 |
120 |
425.87 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.67 |
3.54 |
-67.09 |
2 |
8 |
-1 |
117 |
425.87 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.33 |
Binding ≤ 10μM
|
|
MMP13-2-E |
Matrix Metalloproteinase 13 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.52 |
1.91 |
-16.78 |
3 |
7 |
0 |
105 |
368.386 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.52 |
3.11 |
-53.52 |
2 |
7 |
-1 |
108 |
367.378 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.28 |
Binding ≤ 10μM
|
|
MMP8-4-E |
Matrix Metalloproteinase 8 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.30 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
3.19 |
-14.74 |
0 |
6 |
0 |
70 |
352.39 |
5 |
↓
|
|
|
|
Analogs
-
3961071
-
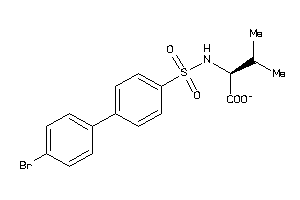
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.30 |
Binding ≤ 10μM
|
|
MMP13-2-E |
Matrix Metalloproteinase 13 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
16 |
0.45 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.48 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.33 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.47 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1500 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
7.48 |
-54.21 |
1 |
5 |
-1 |
86 |
411.297 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.33 |
Binding ≤ 10μM
|
|
MMP13-4-E |
Matrix Metalloproteinase 13 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
11 |
0.46 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.51 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.50 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4500 |
0.31 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3900 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
7.61 |
-46.41 |
1 |
5 |
-1 |
86 |
346.428 |
6 |
↓
|
|
|
|
Analogs
-
13680271
-
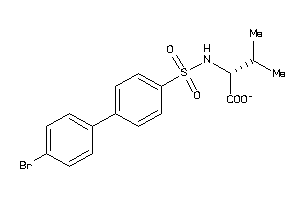
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.30 |
Binding ≤ 10μM
|
|
MMP13-2-E |
Matrix Metalloproteinase 13 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.47 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.33 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.48 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.30 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7900 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
7.65 |
-42.79 |
1 |
5 |
-1 |
86 |
411.297 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
110 |
0.39 |
Binding ≤ 10μM
|
|
MMP13-3-E |
Matrix Metalloproteinase 13 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.45 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
140 |
0.38 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.51 |
3.96 |
-22.07 |
3 |
6 |
0 |
95 |
427.32 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.51 |
5.18 |
-64.2 |
2 |
6 |
-1 |
98 |
426.312 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA17-2-E |
ADAM17 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.32 |
Binding ≤ 10μM
|
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.30 |
Binding ≤ 10μM
|
|
MMP13-2-E |
Matrix Metalloproteinase 13 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.43 |
Binding ≤ 10μM
|
|
MMP14-2-E |
Matrix Metalloproteinase 14 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
14 |
0.39 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.28 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.43 |
Binding ≤ 10μM
|
|
Z102240-1-O |
Cartilage (cluster #1 Of 1), Other |
Other |
30 |
0.38 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.49 |
-5.05 |
-25.61 |
2 |
7 |
0 |
101 |
425.89 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.49 |
-4.49 |
-67.73 |
1 |
7 |
-1 |
104 |
424.882 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8710 |
0.28 |
Binding ≤ 10μM
|
|
MMP13-2-E |
Matrix Metalloproteinase 13 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.46 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.46 |
Binding ≤ 10μM
|
|
MMP3-2-E |
Matrix Metalloproteinase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.45 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.29 |
Binding ≤ 10μM
|
|
MMP8-4-E |
Matrix Metalloproteinase 8 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
11 |
0.45 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
747 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.76 |
6.25 |
-45.85 |
1 |
6 |
-1 |
96 |
362.427 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4540 |
0.22 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
38 |
0.31 |
Binding ≤ 10μM
|
|
MMP14-1-E |
Matrix Metalloproteinase 14 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
91 |
0.29 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.32 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3025 |
0.23 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
8.67 |
-62.63 |
1 |
9 |
-1 |
125 |
509.626 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.30 |
6.19 |
-61.75 |
1 |
9 |
-1 |
129 |
431.446 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA17-2-E |
ADAM17 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Binding ≤ 10μM
|
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
MMP13-3-E |
Matrix Metalloproteinase 13 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.43 |
Binding ≤ 10μM
|
|
MMP14-2-E |
Matrix Metalloproteinase 14 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
91 |
0.35 |
Binding ≤ 10μM
|
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.45 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
8600 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.69 |
4.53 |
-19.46 |
2 |
8 |
0 |
109 |
423.516 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
1.87 |
3.33 |
-11.84 |
2 |
8 |
0 |
112 |
423.516 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.87 |
4.17 |
-47.82 |
1 |
8 |
-1 |
115 |
422.508 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.31 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
41 |
0.30 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
157 |
0.28 |
Binding ≤ 10μM
|
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.46 |
-2.77 |
-15.67 |
3 |
10 |
0 |
127 |
499.678 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.46 |
-3.73 |
-59.76 |
3 |
10 |
-1 |
127 |
498.67 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1210 |
0.27 |
Binding ≤ 10μM
|
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.38 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
44 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.10 |
5.69 |
-52.56 |
3 |
8 |
1 |
106 |
449.549 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.10 |
6.92 |
-70.06 |
2 |
8 |
0 |
109 |
448.541 |
9 |
↓
|
|