|
|
Analogs
-
3993547
-
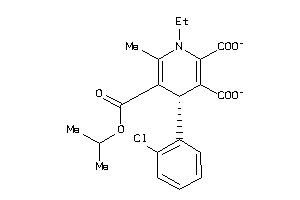
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PYGL-1-E |
Liver Glycogen Phosphorylase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
39 |
0.37 |
Binding ≤ 10μM
|
|
PYGM-1-E |
Muscle Glycogen Phosphorylase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
138 |
0.34 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
2330 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
13.26 |
-138.37 |
0 |
7 |
-2 |
110 |
405.834 |
7 |
↓
|
|
|
|
Analogs
-
3776693
-
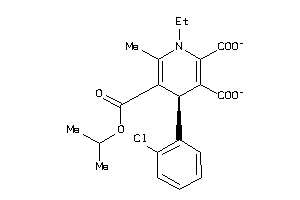
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PYGL-1-E |
Liver Glycogen Phosphorylase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
39 |
0.37 |
Binding ≤ 10μM
|
|
PYGM-1-E |
Muscle Glycogen Phosphorylase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
138 |
0.34 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
2330 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
13.28 |
-138.46 |
0 |
7 |
-2 |
110 |
405.834 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PYGL-1-E |
Liver Glycogen Phosphorylase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.36 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
380 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
4.05 |
-19.03 |
4 |
8 |
0 |
109 |
412.78 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
2585429
-
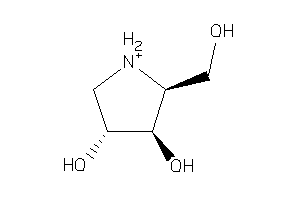
-
15657755
-
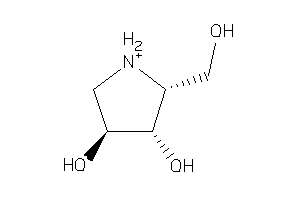
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B1WC34-1-E |
Alpha-glucosidase II Beta Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9700 |
0.78 |
Binding ≤ 10μM
|
|
D3ZAN3-1-E |
Alpha-glucosidase II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9700 |
0.78 |
Binding ≤ 10μM
|
|
GDE-1-E |
Glycogen Debranching Enzyme (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8400 |
0.79 |
Binding ≤ 10μM
|
|
PYGL-2-E |
Liver Glycogen Phosphorylase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
370 |
1.00 |
Binding ≤ 10μM
|
|
PYGM-4-E |
Muscle Glycogen Phosphorylase (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
396 |
1.00 |
Binding ≤ 10μM
|
|
SUIS-1-E |
Sucrase-isomaltase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5800 |
0.81 |
Binding ≤ 10μM
|
|
AMYG-1-F |
Glucoamylase, Intracellular Sporulation-specific (cluster #1 Of 1), Fungal |
Fungi |
150 |
1.06 |
Binding ≤ 10μM
|
|
MAL62-2-F |
Alpha-glucosidase MAL62 (cluster #2 Of 2), Fungal |
Fungi |
840 |
0.95 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.98 |
-7.72 |
-35.36 |
5 |
4 |
1 |
77 |
134.155 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.98 |
-9.1 |
-6.49 |
4 |
4 |
0 |
73 |
133.147 |
1 |
↓
|
|
|
|
Analogs
-
12502895
-
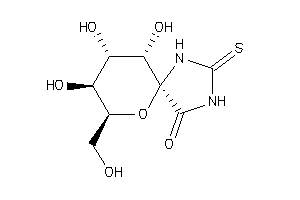
-
12502898
-
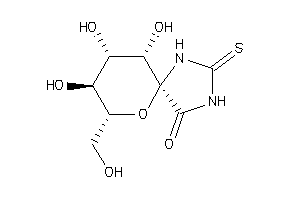
-
3872695
-
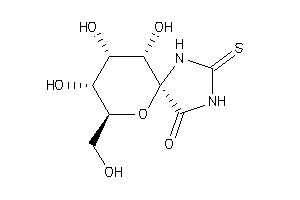
-
3872694
-
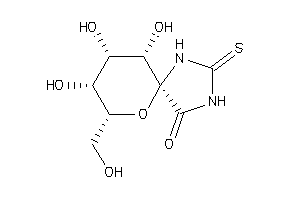
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,3S,4S,5R,6S)-3,4,5-Trihydroxy-2-hydroxymethyl-7,9-diaza-1-oxa-spiro[4,5]decane-10-one-8-thione
(2R,3S,4S,5R,6S)-3,4,5-Trihydrox…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PYGB-1-E |
Brain Glycogen Phosphorylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.42 |
Binding ≤ 10μM
|
|
PYGL-2-E |
Liver Glycogen Phosphorylase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5100 |
0.44 |
Binding ≤ 10μM
|
|
PYGM-4-E |
Muscle Glycogen Phosphorylase (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
5100 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.78 |
-16.4 |
-14.06 |
6 |
8 |
0 |
131 |
264.259 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PYGL-1-E |
Liver Glycogen Phosphorylase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
330 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.64 |
7.18 |
-54.88 |
3 |
6 |
-1 |
105 |
371.8 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PYGL-1-E |
Liver Glycogen Phosphorylase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
130 |
0.34 |
Binding ≤ 10μM
|
|
Z80477-1-O |
SK_HEP1 (Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
1500 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
-2.57 |
-15.46 |
3 |
6 |
0 |
85 |
399.878 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PYGL-1-E |
Liver Glycogen Phosphorylase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
7.74 |
-15.61 |
2 |
5 |
0 |
65 |
335.407 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PYGL-1-E |
Liver Glycogen Phosphorylase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
43 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.34 |
10.96 |
-14.87 |
2 |
4 |
0 |
58 |
486.226 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP51A-1-E |
Cytochrome P450 51 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.24 |
Binding ≤ 10μM
|
|
PYGL-1-E |
Liver Glycogen Phosphorylase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
920 |
0.27 |
Binding ≤ 10μM
|
|
PYGM-2-E |
Muscle Glycogen Phosphorylase (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
-4.39 |
-13.98 |
3 |
6 |
0 |
85 |
443.906 |
5 |
↓
|
|