|
|
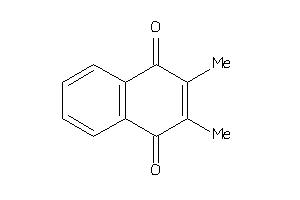
Analogs
-
3063275
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 56 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
5.73 |
-6.36 |
0 |
2 |
0 |
34 |
172.183 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 64 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-1-E |
Androgen Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
26 |
0.53 |
Binding ≤ 10μM
|
|
ERR1-1-E |
Estrogen-related Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
ERR2-1-E |
Estrogen-related Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.59 |
Binding ≤ 10μM
|
|
GPER-1-E |
G-protein Coupled Estrogen Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.58 |
Binding ≤ 10μM
|
|
SHBG-1-E |
Testis-specific Androgen-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.51 |
Binding ≤ 10μM
|
|
ERR1-1-E |
Estrogen-related Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.58 |
Functional ≤ 10μM |
|
ERR2-1-E |
Estrogen-related Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Functional ≤ 10μM |
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.51 |
Functional ≤ 10μM |
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.56 |
Functional ≤ 10μM
|
|
GPER-1-E |
G-protein Coupled Estrogen Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
ADO-2-E |
Aldehyde Oxidase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
ADME/T ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1 |
0.63 |
Functional ≤ 10μM
|
|
Z80226-1-O |
MCF7-2A (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z80475-2-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #2 Of 3), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z81068-1-O |
Ishikawa (Uterine Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
1 |
0.63 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
4.8 |
-5.19 |
2 |
2 |
0 |
40 |
272.388 |
0 |
↓
|
|
|
|
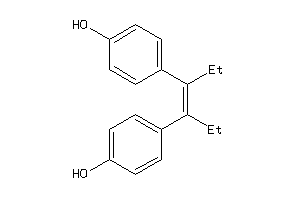
Analogs
-
56544
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 57 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.56 |
Binding ≤ 10μM
|
|
ERR3-1-E |
Estrogen-related Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
630 |
0.43 |
Functional ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
ADO-2-E |
Aldehyde Oxidase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.44 |
ADME/T ≤ 10μM
|
|
Z80005-2-O |
3T3 (Fibroblast Cells) (cluster #2 Of 2), Other |
Other |
2500 |
0.39 |
Functional ≤ 10μM
|
|
Z80030-2-O |
ANN-1 (cluster #2 Of 2), Other |
Other |
3000 |
0.39 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
5000 |
0.37 |
Functional ≤ 10μM
|
|
Z50418-2-O |
Trypanosoma Brucei (cluster #2 Of 3), Other |
Other |
2800 |
0.39 |
ADME/T ≤ 10μM
|
|
Z80092-2-O |
CHO-K1 (Ovarian Cells) (cluster #2 Of 3), Other |
Other |
8500 |
0.35 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
6.1 |
-5.38 |
2 |
2 |
0 |
40 |
268.356 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SCN1A-2-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.29 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.29 |
Binding ≤ 10μM
|
|
SCN3A-2-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.29 |
Binding ≤ 10μM
|
|
SCN8A-2-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.29 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
17 |
0.42 |
Binding ≤ 10μM
|
|
ADO-2-E |
Aldehyde Oxidase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
300 |
0.35 |
ADME/T ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
1100 |
0.32 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
2512 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.65 |
14.31 |
-36.22 |
1 |
3 |
1 |
31 |
354.514 |
11 |
↓
|
|
|
|
|
|
|
|