|
|
Analogs
-
56556
-
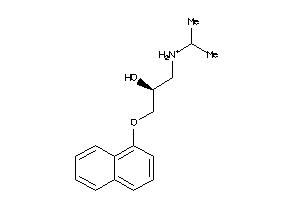
-
4071759
-
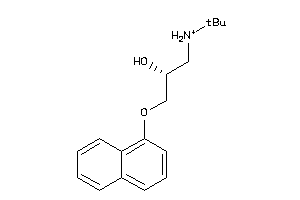
-
32102194
-
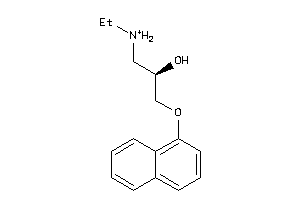
-
32102197
-
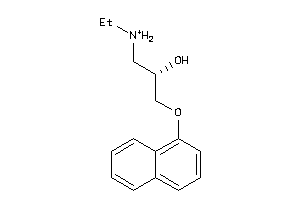
Draw
Identity
99%
90%
80%
70%
Vendors
And 72 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
90 |
0.52 |
Binding ≤ 10μM
|
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1700 |
0.43 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9700 |
0.37 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.60 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.60 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.42 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.47 |
ADME/T ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
3162 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
5.45 |
-42.19 |
3 |
3 |
1 |
46 |
260.357 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.97 |
4.16 |
-6.9 |
2 |
3 |
0 |
41 |
259.349 |
6 |
↓
|
|
|
|
Analogs
-
4071759
-

-
32102194
-

-
32102197
-

-
20240
-
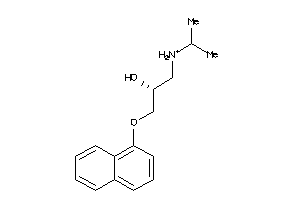
Draw
Identity
99%
90%
80%
70%
Vendors
And 69 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
55 |
0.53 |
Binding ≤ 10μM
|
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
90 |
0.52 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.48 |
Binding ≤ 10μM
|
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.48 |
Binding ≤ 10μM
|
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8100 |
0.38 |
Binding ≤ 10μM
|
|
5HT2B-3-E |
Serotonin 2b (5-HT2b) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.39 |
Binding ≤ 10μM
|
|
5HT2C-3-E |
Serotonin 2c (5-HT2c) Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.43 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.66 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9700 |
0.37 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.66 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.60 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.66 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.60 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN2A-2-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
5HT1B-2-E |
Serotonin 1b (5-HT1b) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4100 |
0.40 |
Functional ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.43 |
ADME/T ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.42 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.48 |
ADME/T ≤ 10μM
|
|
CP2DQ-1-E |
Cytochrome P450 2D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.47 |
ADME/T ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
3162 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
5.47 |
-42.1 |
3 |
3 |
1 |
46 |
260.357 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.97 |
4.28 |
-6.82 |
2 |
3 |
0 |
41 |
259.349 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.00 |
8.67 |
-77.63 |
3 |
5 |
0 |
86 |
363.841 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.35 |
Binding ≤ 10μM |
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2350 |
0.30 |
Binding ≤ 10μM |
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.34 |
Binding ≤ 10μM |
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
550 |
0.34 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.77 |
6.12 |
-83.92 |
3 |
7 |
0 |
105 |
361.394 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.77 |
4.67 |
-52.33 |
2 |
7 |
-1 |
100 |
360.386 |
12 |
↓
|
|
|
|
Analogs
-
25726346
-
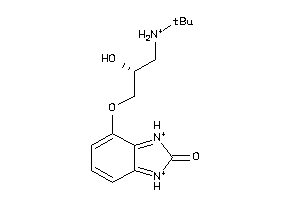
-
34482604
-
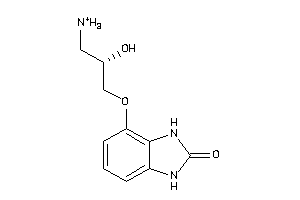
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.40 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.41 |
0.39 |
-47.84 |
5 |
6 |
1 |
95 |
280.348 |
6 |
↓
|
|
|
|
Analogs
-
6021004
-
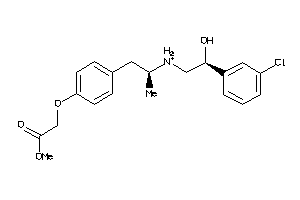
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
59 |
0.39 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.41 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1150 |
0.32 |
Binding ≤ 10μM |
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.34 |
Functional ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
16 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
8.74 |
-52.41 |
3 |
5 |
1 |
72 |
378.876 |
10 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.98 |
3.19 |
-42.74 |
4 |
4 |
1 |
62 |
249.334 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.98 |
2.01 |
-8.94 |
3 |
4 |
0 |
57 |
248.326 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.98 |
3.18 |
-42.7 |
4 |
4 |
1 |
62 |
249.334 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.98 |
1.9 |
-9.07 |
3 |
4 |
0 |
57 |
248.326 |
6 |
↓
|
|
|
|
Analogs
-
4008295
-
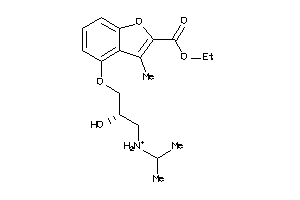
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
98 |
0.41 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
98 |
0.41 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
98 |
0.41 |
Binding ≤ 10μM
|
|
NMT1-1-E |
Peptide N-myristoyltransferase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.35 |
Binding ≤ 10μM
|
|
NMT2-1-E |
Peptide N-myristoyltransferase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
5.78 |
-48.97 |
3 |
6 |
1 |
86 |
336.408 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.91 |
4.57 |
-10.54 |
2 |
6 |
0 |
81 |
335.4 |
9 |
↓
|
|
|
|
Analogs
-
4008294
-
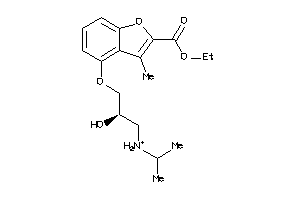
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
98 |
0.41 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.47 |
Binding ≤ 10μM |
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
98 |
0.41 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
98 |
0.41 |
Binding ≤ 10μM
|
|
NMT1-1-E |
Peptide N-myristoyltransferase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.35 |
Binding ≤ 10μM
|
|
NMT2-1-E |
Peptide N-myristoyltransferase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
5.81 |
-48.86 |
3 |
6 |
1 |
86 |
336.408 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.91 |
4.55 |
-10.45 |
2 |
6 |
0 |
81 |
335.4 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.26 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.03 |
4.44 |
-81.56 |
3 |
9 |
0 |
121 |
446.529 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.03 |
2.99 |
-50.59 |
2 |
9 |
-1 |
117 |
445.521 |
12 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.03 |
4.79 |
-103.52 |
5 |
9 |
2 |
121 |
448.545 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.03 |
4.42 |
-81.45 |
3 |
9 |
0 |
121 |
446.529 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.03 |
3.01 |
-50.76 |
2 |
9 |
-1 |
117 |
445.521 |
12 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.03 |
4.77 |
-103.47 |
5 |
9 |
2 |
121 |
448.545 |
12 |
↓
|
|
|
|
Analogs
-
19843876
-
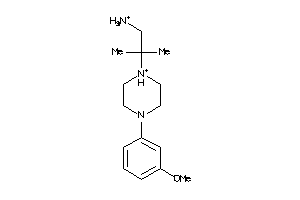
-
34989992
-
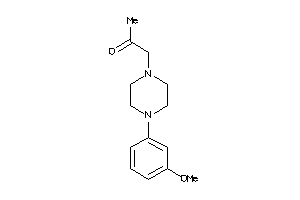
Draw
Identity
99%
90%
80%
70%
Vendors
And 75 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
320 |
0.65 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
2.66 |
-43 |
2 |
3 |
1 |
29 |
193.27 |
2 |
↓
|
|
|
|
Analogs
-
896463
-
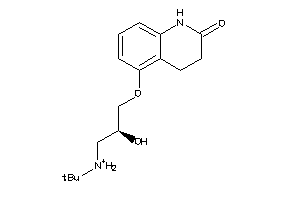
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.96 |
2.8 |
-45.61 |
4 |
5 |
1 |
75 |
293.387 |
6 |
↓
|
|
|
|
Analogs
-
39333910
-
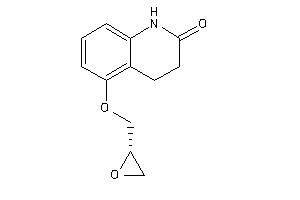
-
39333911
-
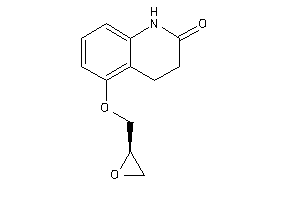
-
128
-
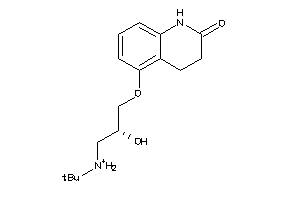
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.96 |
2.82 |
-45.65 |
4 |
5 |
1 |
75 |
293.387 |
6 |
↓
|
|
|
|
Analogs
-
2580783
-
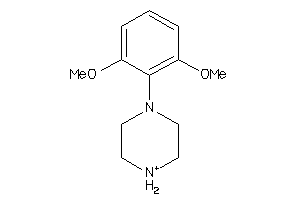
-
34434918
-
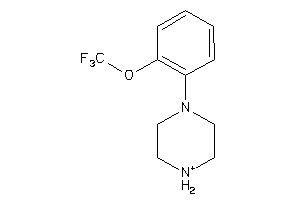
-
36472586
-
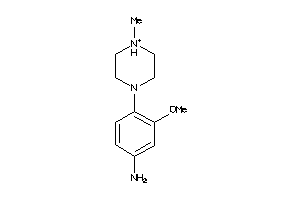
Draw
Identity
99%
90%
80%
70%
Vendors
And 105 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.80 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.75 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.75 |
Binding ≤ 10μM
|
|
5HT1E-2-E |
Serotonin 1e (5-HT1e) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.75 |
Binding ≤ 10μM
|
|
5HT1F-2-E |
Serotonin 1f (5-HT1f) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.75 |
Binding ≤ 10μM
|
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.55 |
Binding ≤ 10μM
|
|
5HT2B-3-E |
Serotonin 2b (5-HT2b) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3500 |
0.55 |
Binding ≤ 10μM
|
|
5HT2C-3-E |
Serotonin 2c (5-HT2c) Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.55 |
Binding ≤ 10μM
|
|
5HT3A-2-E |
Serotonin 3a (5-HT3a) Receptor (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
2500 |
0.56 |
Binding ≤ 10μM
|
|
5HT3B-2-E |
Serotonin 3b (5-HT3b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2500 |
0.56 |
Binding ≤ 10μM
|
|
5HT3C-2-E |
Serotonin 3c (5-HT3c) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.56 |
Binding ≤ 10μM
|
|
5HT3D-2-E |
Serotonin 3d (5-HT3d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.56 |
Binding ≤ 10μM
|
|
5HT3E-2-E |
Serotonin 3e (5-HT3e) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.56 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.90 |
Binding ≤ 10μM
|
|
ADA1A-3-E |
Alpha-1a Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3508 |
0.55 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.57 |
Binding ≤ 10μM
|
|
DRD2-11-E |
Dopamine D2 Receptor (cluster #11 Of 24), Eukaryotic |
Eukaryotes |
1200 |
0.59 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
510 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.35 |
4.29 |
-39.16 |
2 |
3 |
1 |
29 |
193.27 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.35 |
2.98 |
-3.93 |
1 |
3 |
0 |
24 |
192.262 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.40 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
275 |
0.37 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.25 |
3.77 |
-49.25 |
2 |
6 |
1 |
57 |
347.435 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.25 |
2.4 |
-10.91 |
1 |
6 |
0 |
52 |
346.427 |
5 |
↓
|
|
|
|
Analogs
-
1530718
-
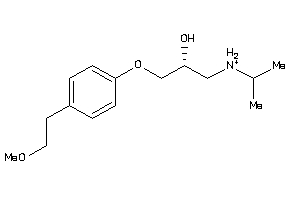
-
70519352
-
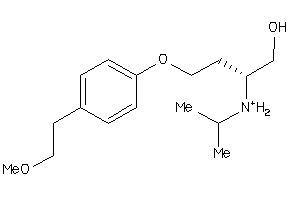
Draw
Identity
99%
90%
80%
70%
Vendors
And 68 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.46 |
Binding ≤ 10μM |
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.46 |
Binding ≤ 10μM |
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.46 |
Binding ≤ 10μM |
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
320 |
0.48 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
3.65 |
-41.5 |
3 |
4 |
1 |
55 |
268.377 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.97 |
2.47 |
-5.96 |
2 |
4 |
0 |
51 |
267.369 |
9 |
↓
|
|
|
|
Analogs
-
70519352
-

-
1530717
-
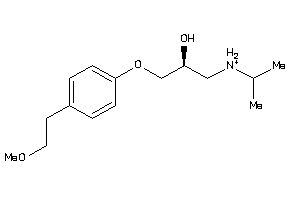
Draw
Identity
99%
90%
80%
70%
Vendors
And 62 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.46 |
Binding ≤ 10μM |
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.46 |
Binding ≤ 10μM |
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.46 |
Binding ≤ 10μM |
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
320 |
0.48 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
3.63 |
-41.68 |
3 |
4 |
1 |
55 |
268.377 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.97 |
2.34 |
-6.05 |
2 |
4 |
0 |
51 |
267.369 |
9 |
↓
|
|
|
|
Analogs
-
34962480
-
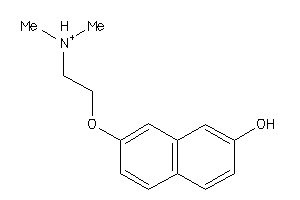
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.62 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.51 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.47 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.47 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.47 |
Binding ≤ 10μM
|
|
5HT1B-2-E |
Serotonin 1b (5-HT1b) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
94 |
0.61 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
2.12 |
-36.57 |
1 |
2 |
1 |
13 |
216.304 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.71 |
-4.63 |
-53.65 |
5 |
6 |
1 |
90 |
481.455 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.71 |
-4.59 |
-53.54 |
5 |
6 |
1 |
90 |
481.455 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.41 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.41 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.71 |
-5.14 |
-56.29 |
5 |
6 |
1 |
90 |
481.455 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.41 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.41 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.71 |
5.89 |
-55.8 |
5 |
6 |
1 |
91 |
481.455 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
77 |
0.33 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.58 |
6.16 |
-51.35 |
3 |
7 |
1 |
90 |
412.51 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
77 |
0.33 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.58 |
6.15 |
-51.34 |
3 |
7 |
1 |
90 |
412.51 |
8 |
↓
|
|
|
|
Analogs
-
86599
-
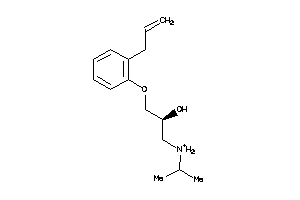
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
5.16 |
-40.25 |
3 |
3 |
1 |
46 |
250.362 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.58 |
3.88 |
-5.22 |
2 |
3 |
0 |
41 |
249.354 |
8 |
↓
|
|
|
|
Analogs
-
3647816
-
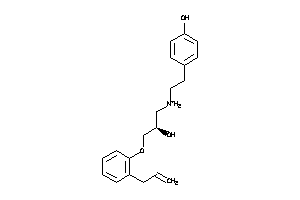
-
3647817
-
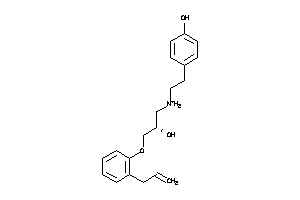
-
23
-
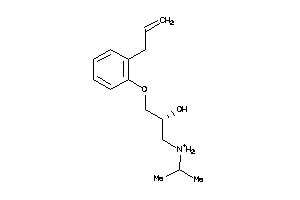
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
5.19 |
-40.14 |
3 |
3 |
1 |
46 |
250.362 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.58 |
4.01 |
-5.14 |
2 |
3 |
0 |
41 |
249.354 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 45 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
80 |
0.66 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
110 |
0.65 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
325 |
0.61 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.81 |
Functional ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.81 |
Functional ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.81 |
Functional ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.56 |
Functional ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.76 |
Functional ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.76 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
7943 |
0.48 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
6 |
0.77 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
226 |
0.62 |
Functional ≤ 10μM
|
|
Z80088-1-O |
CHO (Ovarian Cells) (cluster #1 Of 4), Other |
Other |
1 |
0.84 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.62 |
-1.43 |
-43.02 |
5 |
4 |
1 |
77 |
212.269 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.62 |
-2.62 |
-7.01 |
4 |
4 |
0 |
73 |
211.261 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 50 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
80 |
0.66 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
110 |
0.65 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
325 |
0.61 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.81 |
Functional ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.81 |
Functional ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.81 |
Functional ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.56 |
Functional ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.76 |
Functional ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.76 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
7943 |
0.48 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
10000 |
0.47 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
6 |
0.77 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
226 |
0.62 |
Functional ≤ 10μM
|
|
Z80088-1-O |
CHO (Ovarian Cells) (cluster #1 Of 4), Other |
Other |
1 |
0.84 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.62 |
-1.49 |
-44.52 |
5 |
4 |
1 |
77 |
212.269 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.62 |
-2.75 |
-7.53 |
4 |
4 |
0 |
73 |
211.261 |
4 |
↓
|
|
|
|
Analogs
-
37100949
-
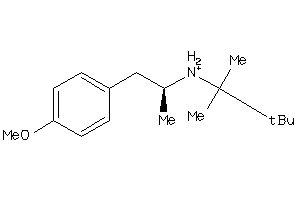
-
37100950
-
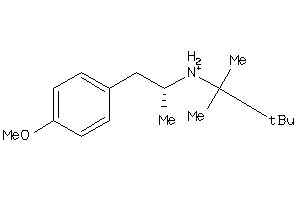
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4940 |
0.35 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4940 |
0.35 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4940 |
0.35 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
10 |
0.53 |
Binding ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
9320 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.61 |
1.16 |
-40.6 |
2 |
2 |
1 |
26 |
284.423 |
8 |
↓
|
|
|
|
Analogs
-
37100949
-

-
37100950
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4940 |
0.35 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4940 |
0.35 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4940 |
0.35 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
10 |
0.53 |
Binding ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
9320 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.61 |
1.16 |
-41.12 |
2 |
2 |
1 |
26 |
284.423 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.45 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.45 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
-1.73 |
-51.8 |
3 |
5 |
1 |
76 |
354.426 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.45 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.45 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
-1.78 |
-51.85 |
3 |
5 |
1 |
76 |
354.426 |
8 |
↓
|
|
|
|
Analogs
-
6091848
-
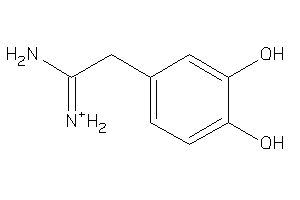
-
36489899
-
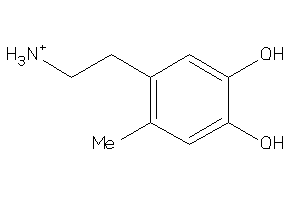
Draw
Identity
99%
90%
80%
70%
Vendors
And 67 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
61 |
0.92 |
Binding ≤ 10μM
|
|
ADA2A-3-E |
Alpha-2a Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
61 |
0.92 |
Binding ≤ 10μM
|
|
ADA2B-4-E |
Alpha-2b Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
61 |
0.92 |
Binding ≤ 10μM
|
|
ADA2C-3-E |
Alpha-2c Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
61 |
0.92 |
Binding ≤ 10μM
|
|
ADCY2-2-E |
Adenylate Cyclase Type II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.67 |
Binding ≤ 10μM
|
|
ADCY3-2-E |
Adenylate Cyclase Type III (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.67 |
Binding ≤ 10μM
|
|
ADCY4-2-E |
Adenylate Cyclase Type IV (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.67 |
Binding ≤ 10μM
|
|
ADCY5-2-E |
Adenylate Cyclase Type V (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.67 |
Binding ≤ 10μM
|
|
ADCY6-2-E |
Adenylate Cyclase Type VI (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.67 |
Binding ≤ 10μM
|
|
ADCY8-1-E |
Adenylate Cyclase Type VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.67 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.64 |
Binding ≤ 10μM
|
|
D4A3N4-1-E |
Brain Adenylate Cyclase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.69 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9 |
1.02 |
Binding ≤ 10μM
|
|
DRD4-2-E |
Dopamine D4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5340 |
0.67 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5340 |
0.67 |
Binding ≤ 10μM
|
|
MTR1A-2-E |
Melatonin Receptor 1A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7100 |
0.66 |
Binding ≤ 10μM |
|
MTR1B-2-E |
Melatonin Receptor 1B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9100 |
0.64 |
Binding ≤ 10μM |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3045 |
0.70 |
Binding ≤ 10μM
|
|
OPRK-2-E |
Kappa Opioid Receptor (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3045 |
0.70 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3045 |
0.70 |
Binding ≤ 10μM
|
|
Q8CFM9-1-E |
Adenylate Cyclase Type VII (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.69 |
Binding ≤ 10μM
|
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
SC6A3-3-E |
Dopamine Transporter (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6400 |
0.66 |
Binding ≤ 10μM
|
|
SGMR1-4-E |
Sigma Opioid Receptor (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
3045 |
0.70 |
Binding ≤ 10μM
|
|
ARP19-1-E |
Cyclic AMP Phosphoprotein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.69 |
Functional ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
35 |
0.95 |
Functional ≤ 10μM
|
|
DRD2-1-E |
Dopamine D2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
1.06 |
Functional ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
1.11 |
Functional ≤ 10μM
|
|
DRD2-3-E |
Dopamine D2 Receptor (cluster #3 Of 24), Eukaryotic |
Eukaryotes |
843 |
0.77 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 5), Other |
Other |
29 |
0.96 |
Binding ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
10000 |
0.64 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
1700 |
0.73 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
76 |
0.91 |
Functional ≤ 10μM
|
|
Z50594-6-O |
Mus Musculus (cluster #6 Of 9), Other |
Other |
10000 |
0.64 |
Functional ≤ 10μM
|
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
5268 |
0.67 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.05 |
-1.62 |
-50.24 |
5 |
3 |
1 |
68 |
154.189 |
2 |
↓
|
|
|
|
Analogs
-
1530580
-
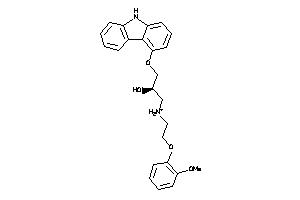
Draw
Identity
99%
90%
80%
70%
Vendors
And 41 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
7.38 |
-51.26 |
4 |
6 |
1 |
80 |
407.49 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.56 |
6.05 |
-14.85 |
3 |
6 |
0 |
76 |
406.482 |
10 |
↓
|
|
|
|
Analogs
-
1530579
-
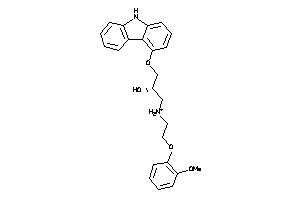
Draw
Identity
99%
90%
80%
70%
Vendors
And 41 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
7.36 |
-51.12 |
4 |
6 |
1 |
80 |
407.49 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.56 |
5.94 |
-15.03 |
3 |
6 |
0 |
76 |
406.482 |
10 |
↓
|
|
|
|
Analogs
-
2382386
-
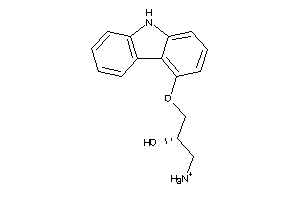
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
-3.65 |
-44.34 |
4 |
4 |
1 |
61 |
299.394 |
6 |
↓
|
|
|
|
Analogs
-
2382386
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
-3.7 |
-44.42 |
4 |
4 |
1 |
61 |
299.394 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
740 |
0.23 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.27 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1480 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
10.99 |
-42.03 |
2 |
7 |
1 |
76 |
502.635 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.01 |
8.66 |
-15.47 |
1 |
7 |
0 |
75 |
501.627 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.01 |
11.01 |
-48.61 |
2 |
7 |
1 |
76 |
502.635 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
740 |
0.23 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.27 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1480 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
10.86 |
-42.24 |
2 |
7 |
1 |
76 |
502.635 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.01 |
8.49 |
-15.69 |
1 |
7 |
0 |
75 |
501.627 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.01 |
10.88 |
-48.6 |
2 |
7 |
1 |
76 |
502.635 |
9 |
↓
|
|
|
|
Analogs
-
359025
-
-
359026
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
114 |
0.49 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.57 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4960 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.00 |
3.39 |
-6.68 |
1 |
4 |
0 |
42 |
277.364 |
7 |
↓
|
|
|
|
Analogs
-
359025
-

-
359026
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
114 |
0.49 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.57 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4960 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.00 |
3.32 |
-6.59 |
1 |
4 |
0 |
42 |
277.364 |
7 |
↓
|
|
|
|
Analogs
-
5051180
-
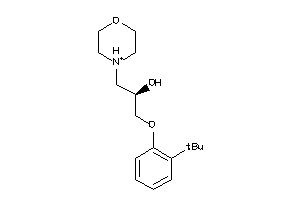
-
5051181
-
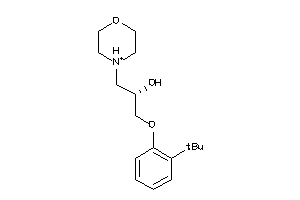
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1770 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.22 |
3.29 |
-6.58 |
1 |
4 |
0 |
42 |
279.38 |
6 |
↓
|
|
|
|
Analogs
-
5051180
-

-
5051181
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1770 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.22 |
3.36 |
-6.6 |
1 |
4 |
0 |
42 |
279.38 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
293 |
0.25 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.26 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3100 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.79 |
10.72 |
-39.34 |
2 |
7 |
1 |
76 |
500.619 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.79 |
10.7 |
-46.61 |
2 |
7 |
1 |
76 |
500.619 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.79 |
8.46 |
-16.6 |
1 |
7 |
0 |
75 |
499.611 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
293 |
0.25 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.26 |
Binding ≤ 10μM
|
|
ADRB3-2-E |
Beta-3 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3100 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.79 |
10.66 |
-38.86 |
2 |
7 |
1 |
76 |
500.619 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.79 |
10.69 |
-46.39 |
2 |
7 |
1 |
76 |
500.619 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.79 |
8.4 |
-16.18 |
1 |
7 |
0 |
75 |
499.611 |
10 |
↓
|
|
|
|
Analogs
-
12170472
-
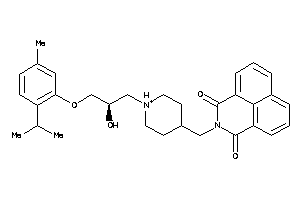
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.31 |
-0.03 |
-44.59 |
2 |
6 |
1 |
72 |
501.647 |
8 |
↓
|
|