|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
-0.86 |
-42.33 |
1 |
7 |
1 |
70 |
360.482 |
6 |
↓
|
|
|
|
Analogs
-
1851395
-
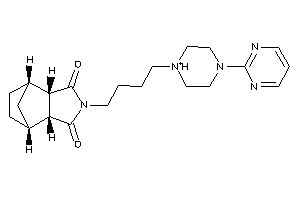
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
25 |
0.38 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
1260 |
0.29 |
Binding ≤ 10μM
|
|
DRD2-16-E |
Dopamine D2 Receptor (cluster #16 Of 24), Eukaryotic |
Eukaryotes |
460 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.74 |
12.37 |
-42.59 |
1 |
7 |
1 |
71 |
384.504 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.74 |
10.15 |
-11.84 |
0 |
7 |
0 |
70 |
383.496 |
6 |
↓
|
|
|
|
Analogs
-
643032
-
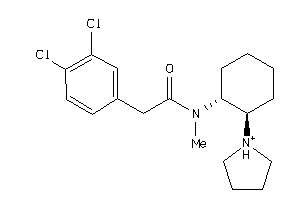
-
591998
-
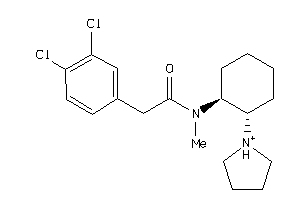
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.57 |
12.11 |
-43.68 |
1 |
3 |
1 |
25 |
370.344 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.47 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.37 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
73 |
0.37 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.37 |
Binding ≤ 10μM
|
|
DRD2-16-E |
Dopamine D2 Receptor (cluster #16 Of 24), Eukaryotic |
Eukaryotes |
166 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.02 |
8.8 |
-11.84 |
1 |
6 |
0 |
61 |
369.513 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
8.34 |
-10.11 |
1 |
5 |
0 |
45 |
397.563 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
25 |
0.41 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
26 |
0.41 |
Binding ≤ 10μM
|
|
DRD2-16-E |
Dopamine D2 Receptor (cluster #16 Of 24), Eukaryotic |
Eukaryotes |
416 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
9.8 |
-45.11 |
1 |
6 |
1 |
48 |
357.478 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.97 |
7.58 |
-11.11 |
0 |
6 |
0 |
47 |
356.47 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
25 |
0.41 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
26 |
0.41 |
Binding ≤ 10μM
|
|
DRD2-16-E |
Dopamine D2 Receptor (cluster #16 Of 24), Eukaryotic |
Eukaryotes |
416 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
9.73 |
-45.11 |
1 |
6 |
1 |
48 |
357.478 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.97 |
7.51 |
-11.45 |
0 |
6 |
0 |
47 |
356.47 |
6 |
↓
|
|
|
|
Analogs
-
6092059
-
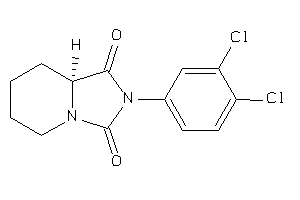
-
6092061
-
-
6092370
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.40 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
12 |
0.40 |
Binding ≤ 10μM |
|
DRD2-16-E |
Dopamine D2 Receptor (cluster #16 Of 24), Eukaryotic |
Eukaryotes |
129 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.13 |
11.62 |
-45.76 |
1 |
6 |
1 |
48 |
405.95 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.13 |
9.4 |
-9.89 |
0 |
6 |
0 |
47 |
404.942 |
6 |
↓
|
|
|
|
Analogs
-
6092059
-

-
6092061
-

-
6092370
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.40 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
12 |
0.40 |
Binding ≤ 10μM |
|
DRD2-16-E |
Dopamine D2 Receptor (cluster #16 Of 24), Eukaryotic |
Eukaryotes |
129 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.13 |
11.61 |
-45.64 |
1 |
6 |
1 |
48 |
405.95 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.13 |
9.39 |
-10.04 |
0 |
6 |
0 |
47 |
404.942 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.36 |
Binding ≤ 10μM |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.32 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
72 |
0.32 |
Binding ≤ 10μM |
|
DRD2-16-E |
Dopamine D2 Receptor (cluster #16 Of 24), Eukaryotic |
Eukaryotes |
91 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.35 |
0.21 |
-46.85 |
1 |
6 |
1 |
48 |
439.502 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.36 |
Binding ≤ 10μM |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.32 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
72 |
0.32 |
Binding ≤ 10μM |
|
DRD2-16-E |
Dopamine D2 Receptor (cluster #16 Of 24), Eukaryotic |
Eukaryotes |
91 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.35 |
11.07 |
-46.55 |
1 |
6 |
1 |
48 |
439.502 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.36 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
10 |
0.40 |
Binding ≤ 10μM
|
|
DRD2-16-E |
Dopamine D2 Receptor (cluster #16 Of 24), Eukaryotic |
Eukaryotes |
615 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.64 |
11.17 |
-47.82 |
1 |
6 |
1 |
48 |
389.495 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.64 |
8.96 |
-10.51 |
0 |
6 |
0 |
47 |
388.487 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.36 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
10 |
0.40 |
Binding ≤ 10μM
|
|
DRD2-16-E |
Dopamine D2 Receptor (cluster #16 Of 24), Eukaryotic |
Eukaryotes |
615 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.64 |
11.16 |
-47.69 |
1 |
6 |
1 |
48 |
389.495 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.64 |
8.95 |
-10.58 |
0 |
6 |
0 |
47 |
388.487 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
11 |
0.41 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.41 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
10 |
0.41 |
Binding ≤ 10μM
|
|
DRD2-16-E |
Dopamine D2 Receptor (cluster #16 Of 24), Eukaryotic |
Eukaryotes |
151 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
10.31 |
-46.95 |
1 |
6 |
1 |
48 |
391.923 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.62 |
8.09 |
-11.24 |
0 |
6 |
0 |
47 |
390.915 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
11 |
0.41 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.41 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
10 |
0.41 |
Binding ≤ 10μM
|
|
DRD2-16-E |
Dopamine D2 Receptor (cluster #16 Of 24), Eukaryotic |
Eukaryotes |
151 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
10.23 |
-47.01 |
1 |
6 |
1 |
48 |
391.923 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.62 |
8.02 |
-11.47 |
0 |
6 |
0 |
47 |
390.915 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.50 |
8.23 |
-10.83 |
0 |
6 |
0 |
53 |
385.508 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 42 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
17 |
0.39 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
95 |
0.35 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
95 |
0.35 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
178 |
0.34 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1250 |
0.30 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2270 |
0.28 |
Binding ≤ 10μM
|
|
ADA1A-2-E |
Alpha-1a Adrenergic Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1320 |
0.29 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1320 |
0.29 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1320 |
0.29 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.25 |
Binding ≤ 10μM
|
|
ADA2B-2-E |
Alpha-2b Adrenergic Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.25 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.25 |
Binding ≤ 10μM
|
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6100 |
0.26 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.31 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
120 |
0.35 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.35 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3981 |
0.27 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
48 |
0.37 |
Binding ≤ 10μM
|
|
5HT1A-2-E |
Serotonin 1a (5-HT1a) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Functional ≤ 10μM
|
|
DRD2-16-E |
Dopamine D2 Receptor (cluster #16 Of 24), Eukaryotic |
Eukaryotes |
852 |
0.30 |
Binding ≤ 10μM |
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
4160 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
12.45 |
-41.93 |
1 |
7 |
1 |
71 |
386.52 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.75 |
10.24 |
-12.76 |
0 |
7 |
0 |
70 |
385.512 |
6 |
↓
|
|
|
|
|