|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5600 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.81 |
2.1 |
-15.07 |
2 |
4 |
0 |
88 |
200.197 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.37 |
Binding ≤ 10μM |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
130 |
0.40 |
Functional ≤ 10μM
|
|
ERBB2-1-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
130 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.64 |
-1.87 |
-11.25 |
3 |
5 |
0 |
96 |
340.404 |
6 |
↓
|
|
|
|
Analogs
-
14640373
-
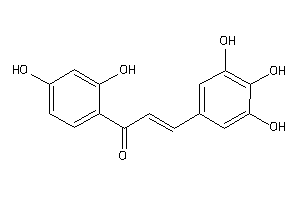
-
4252698
-
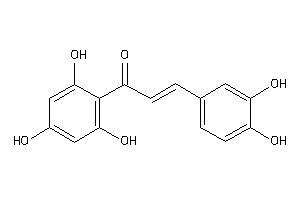
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACE-1-E |
Angiotensin-converting Enzyme (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
730 |
0.43 |
Binding ≤ 10μM
|
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.36 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
Q965D5-1-E |
Enoyl-acyl-carrier Protein Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
0.03 |
-14.86 |
4 |
5 |
0 |
98 |
272.256 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.28 |
0.8 |
-48.48 |
3 |
5 |
-1 |
101 |
271.248 |
3 |
↓
|
|
|
|
Analogs
-
37130512
-
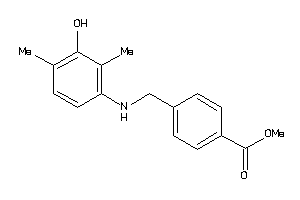
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.45 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.45 |
Binding ≤ 10μM
|
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
-4.45 |
-11.96 |
4 |
6 |
0 |
99 |
289.287 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2380 |
0.39 |
Binding ≤ 10μM |
|
Z81020-4-O |
HepG2 (Hepatoblastoma Cells) (cluster #4 Of 8), Other |
Other |
1460 |
0.41 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.77 |
9.41 |
-11.48 |
3 |
3 |
0 |
50 |
281.384 |
5 |
↓
|
|
|
|
Analogs
-
6484558
-
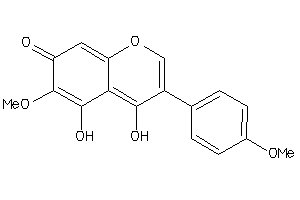
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3300 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
-2.06 |
-16.8 |
3 |
6 |
0 |
100 |
300.266 |
2 |
↓
|
|
|
|
Analogs
-
160657
-
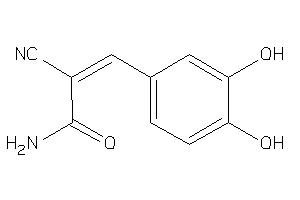
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4500 |
0.50 |
Binding ≤ 10μM
|
|
ERBB2-1-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6400 |
0.48 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7000 |
0.48 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.15 |
-2.32 |
-12.35 |
4 |
5 |
0 |
107 |
204.185 |
2 |
↓
|
|
|
|
Analogs
-
8617556
-
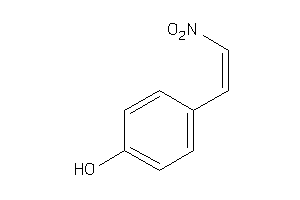
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.63 |
3.72 |
-8 |
1 |
4 |
0 |
66 |
165.148 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.51 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.44 |
3.76 |
-9.14 |
1 |
5 |
0 |
75 |
195.174 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.45 |
4.68 |
-41.97 |
0 |
5 |
-1 |
78 |
194.166 |
3 |
↓
|
|
|
|
Analogs
-
34541434
-
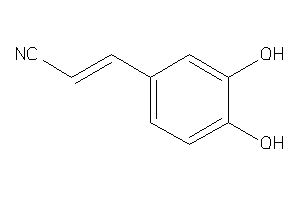
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.52 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.52 |
Binding ≤ 10μM
|
|
LUXP-1-B |
Autoinducer 2-binding Periplasmic Protein LuxP (cluster #1 Of 2), Bacterial |
Bacteria |
3000 |
0.52 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
6310 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.91 |
-0.8 |
-14.91 |
3 |
5 |
0 |
108 |
202.169 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.91 |
0.23 |
-47.75 |
2 |
5 |
-1 |
111 |
201.161 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.91 |
0.11 |
-52.62 |
2 |
5 |
-1 |
111 |
201.161 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5900 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.43 |
-2.84 |
-11.45 |
3 |
5 |
0 |
93 |
286.331 |
3 |
↓
|
|
|
|
Analogs
-
21288966
-
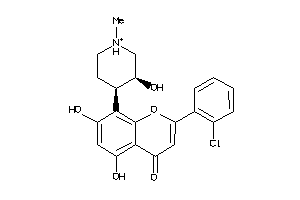
-
6145162
-
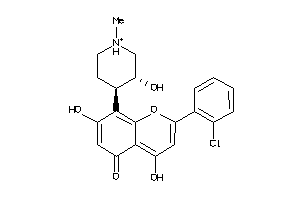
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.33 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
220 |
0.33 |
Binding ≤ 10μM
|
|
CDK4-2-E |
Cyclin-dependent Kinase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.32 |
Binding ≤ 10μM
|
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4600 |
0.27 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.32 |
Binding ≤ 10μM
|
|
GSK3B-7-E |
Glycogen Synthase Kinase-3 Beta (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
450 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.46 |
6.36 |
-53.64 |
4 |
6 |
1 |
95 |
402.854 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.46 |
7.12 |
-63.32 |
3 |
6 |
0 |
98 |
401.846 |
2 |
↓
|
|
|
|
Analogs
-
29416077
-
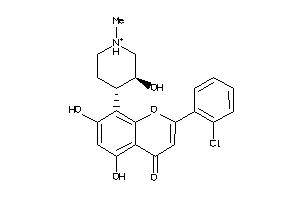
-
18825325
-
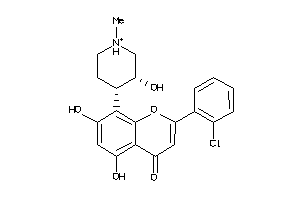
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AAK1-1-E |
Adaptor-associated Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5300 |
0.26 |
Binding ≤ 10μM
|
|
AAPK1-1-E |
AMP-activated Protein Kinase, Alpha-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6600 |
0.26 |
Binding ≤ 10μM
|
|
ACVR1-1-E |
Activin Receptor Type-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.29 |
Binding ≤ 10μM
|
|
ALK-1-E |
ALK Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
670 |
0.31 |
Binding ≤ 10μM
|
|
AVR2B-1-E |
Activin Receptor Type-2B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2400 |
0.28 |
Binding ≤ 10μM
|
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
70 |
0.36 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
70 |
0.36 |
Binding ≤ 10μM
|
|
CCNB-1-E |
G2/mitotic-specific Cyclin B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.32 |
Binding ≤ 10μM |
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.31 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.31 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.31 |
Binding ≤ 10μM
|
|
CCND1-1-E |
G1/S-specific Cyclin D1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
CCND2-1-E |
G1/S-specific Cyclin D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
CCND3-1-E |
G1/S-specific Cyclin D3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
CCNE1-1-E |
G1/S-specific Cyclin E1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
960 |
0.30 |
Binding ≤ 10μM
|
|
CCNE2-1-E |
G1/S-specific Cyclin E2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
960 |
0.30 |
Binding ≤ 10μM
|
|
CCNH-1-E |
Cyclin H (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.32 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
520 |
0.31 |
Binding ≤ 10μM
|
|
CDK14-1-E |
Serine/threonine-protein Kinase PFTAIRE-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.35 |
Binding ≤ 10μM
|
|
CDK16-1-E |
Serine/threonine-protein Kinase PCTAIRE-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
440 |
0.32 |
Binding ≤ 10μM
|
|
CDK17-1-E |
Serine/threonine-protein Kinase PCTAIRE-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
480 |
0.32 |
Binding ≤ 10μM
|
|
CDK18-1-E |
Serine/threonine-protein Kinase PCTAIRE-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.30 |
Binding ≤ 10μM
|
|
CDK19-1-E |
Cell Division Cycle 2-like Protein Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
57 |
0.36 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
960 |
0.30 |
Binding ≤ 10μM
|
|
CDK3-1-E |
Cyclin-dependent Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
410 |
0.32 |
Binding ≤ 10μM
|
|
CDK4-2-E |
Cyclin-dependent Kinase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
CDK5-1-E |
Cyclin-dependent Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
43 |
0.37 |
Binding ≤ 10μM
|
|
CDK6-1-E |
Cyclin-dependent Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.35 |
Binding ≤ 10μM
|
|
CDK7-1-E |
Cyclin-dependent Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
875 |
0.30 |
Binding ≤ 10μM
|
|
CDK8-1-E |
Cell Division Protein Kinase 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.35 |
Binding ≤ 10μM
|
|
CDK9-1-E |
Cyclin-dependent Kinase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Binding ≤ 10μM
|
|
CLK1-1-E |
Dual Specificty Protein Kinase CLK1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.28 |
Binding ≤ 10μM
|
|
CLK2-1-E |
Dual Specificity Protein Kinase CLK2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.28 |
Binding ≤ 10μM
|
|
CLK3-1-E |
Dual Specificity Protein Kinase CLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.29 |
Binding ≤ 10μM
|
|
CSF1R-1-E |
Macrophage Colony-stimulating Factor 1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.28 |
Binding ≤ 10μM
|
|
CTRO-1-E |
Citron Rho-interacting Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.35 |
Binding ≤ 10μM
|
|
DCLK1-1-E |
Serine/threonine-protein Kinase DCLK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.28 |
Binding ≤ 10μM
|
|
DCLK3-1-E |
Serine/threonine-protein Kinase DCLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.30 |
Binding ≤ 10μM
|
|
DMPK-1-E |
Myotonin-protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
650 |
0.31 |
Binding ≤ 10μM
|
|
DYR1B-1-E |
Dual Specificity Tyrosine-phosphorylation-regulated Kinase 1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
84 |
0.35 |
Binding ≤ 10μM
|
|
E2AK2-1-E |
Interferon-induced, Double-stranded RNA-activated Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5200 |
0.26 |
Binding ≤ 10μM
|
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7600 |
0.26 |
Binding ≤ 10μM
|
|
EPHA2-1-E |
Ephrin Type-A Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.28 |
Binding ≤ 10μM
|
|
EPHA3-1-E |
Ephrin Type-A Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
EPHA5-1-E |
Ephrin Type-A Receptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.27 |
Binding ≤ 10μM
|
|
EPHB1-1-E |
Ephrin Type-B Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.28 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6500 |
0.26 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
370 |
0.32 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1300 |
0.29 |
Binding ≤ 10μM
|
|
GSK3B-7-E |
Glycogen Synthase Kinase-3 Beta (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
730 |
0.31 |
Binding ≤ 10μM
|
|
IKKE-1-E |
Inhibitor Of Nuclear Factor Kappa B Kinase Epsilon Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.28 |
Binding ≤ 10μM
|
|
KCC2A-1-E |
CaM Kinase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.29 |
Binding ≤ 10μM
|
|
KCC2B-1-E |
CaM Kinase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.28 |
Binding ≤ 10μM
|
|
KCC4-1-E |
CaM Kinase IV (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3200 |
0.27 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4600 |
0.27 |
Binding ≤ 10μM
|
|
KKCC1-1-E |
CaM-kinase Kinase Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
79 |
0.36 |
Binding ≤ 10μM
|
|
KKCC2-1-E |
CaM-kinase Kinase Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
430 |
0.32 |
Binding ≤ 10μM
|
|
KPCA-1-E |
Protein Kinase C Alpha (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2800 |
0.28 |
Binding ≤ 10μM
|
|
KPCB-1-E |
Protein Kinase C Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.25 |
Binding ≤ 10μM
|
|
KPCD-1-E |
Protein Kinase C Delta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
890 |
0.30 |
Binding ≤ 10μM
|
|
KPCD1-1-E |
Protein Kinase C Mu (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
520 |
0.31 |
Binding ≤ 10μM
|
|
KPCD2-1-E |
Serine/threonine-protein Kinase D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.30 |
Binding ≤ 10μM
|
|
KPCD3-1-E |
Protein Kinase C Nu (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
170 |
0.34 |
Binding ≤ 10μM
|
|
KPCE-1-E |
Protein Kinase C Epsilon (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
480 |
0.32 |
Binding ≤ 10μM
|
|
KPCG-1-E |
Protein Kinase C Gamma (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.25 |
Binding ≤ 10μM
|
|
KPCI-1-E |
Protein Kinase C Iota (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.25 |
Binding ≤ 10μM
|
|
KPCL-2-E |
Protein Kinase C Eta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
350 |
0.32 |
Binding ≤ 10μM
|
|
KPCT-1-E |
Protein Kinase C Theta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
350 |
0.32 |
Binding ≤ 10μM
|
|
KPCZ-1-E |
Protein Kinase C Zeta (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10000 |
0.25 |
Binding ≤ 10μM
|
|
KS6A1-1-E |
Ribosomal Protein S6 Kinase Alpha 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
720 |
0.31 |
Binding ≤ 10μM
|
|
KS6A2-1-E |
Ribosomal Protein S6 Kinase Alpha 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.29 |
Binding ≤ 10μM
|
|
KS6A4-1-E |
Ribosomal Protein S6 Kinase Alpha 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.29 |
Binding ≤ 10μM
|
|
KS6A5-1-E |
Ribosomal Protein S6 Kinase Alpha 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.28 |
Binding ≤ 10μM
|
|
KS6A6-1-E |
Ribosomal Protein S6 Kinase Alpha 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.30 |
Binding ≤ 10μM
|
|
LTK-1-E |
Leukocyte Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.28 |
Binding ≤ 10μM
|
|
M3K10-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5200 |
0.26 |
Binding ≤ 10μM
|
|
M3K9-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 9 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5700 |
0.26 |
Binding ≤ 10μM
|
|
M4K1-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
M4K3-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4500 |
0.27 |
Binding ≤ 10μM
|
|
MERTK-1-E |
Proto-oncogene Tyrosine-protein Kinase MER (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6800 |
0.26 |
Binding ≤ 10μM
|
|
MK07-1-E |
Mitogen-activated Protein Kinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
620 |
0.31 |
Binding ≤ 10μM
|
|
MK10-1-E |
C-Jun N-terminal Kinase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5500 |
0.26 |
Binding ≤ 10μM
|
|
MK15-1-E |
Mitogen-activated Protein Kinase 15 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
330 |
0.32 |
Binding ≤ 10μM
|
|
MKNK2-1-E |
MAP Kinase Signal-integrating Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.27 |
Binding ≤ 10μM
|
|
MRCKA-1-E |
Serine/threonine-protein Kinase MRCK-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9500 |
0.25 |
Binding ≤ 10μM
|
|
MRCKB-1-E |
Serine/threonine-protein Kinase MRCK Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.27 |
Binding ≤ 10μM
|
|
NUAK2-1-E |
NUAK Family SNF1-like Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.29 |
Binding ≤ 10μM
|
|
PHKG1-1-E |
Phosphorylase Kinase Gamma Subunit 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.28 |
Binding ≤ 10μM
|
|
PHKG2-1-E |
Phosphorylase Kinase Gamma Subunit 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2900 |
0.28 |
Binding ≤ 10μM
|
|
PIM1-1-E |
Serine/threonine-protein Kinase PIM1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
560 |
0.31 |
Binding ≤ 10μM
|
|
PIM2-1-E |
Serine/threonine-protein Kinase PIM2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
770 |
0.31 |
Binding ≤ 10μM
|
|
PIM3-1-E |
Serine/threonine-protein Kinase PIM3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.31 |
Binding ≤ 10μM
|
|
PKN1-1-E |
Protein Kinase N1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.27 |
Binding ≤ 10μM
|
|
PLK4-1-E |
Serine/threonine-protein Kinase PLK4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.28 |
Binding ≤ 10μM
|
|
PYGM-2-E |
Muscle Glycogen Phosphorylase (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Binding ≤ 10μM
|
|
SRMS-1-E |
Tyrosine-protein Kinase Srms (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.29 |
Binding ≤ 10μM
|
|
ST17B-1-E |
Serine/threonine-protein Kinase 17B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5100 |
0.26 |
Binding ≤ 10μM
|
|
STK3-1-E |
Serine/threonine-protein Kinase MST2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9400 |
0.25 |
Binding ≤ 10μM
|
|
STK33-1-E |
Serine/threonine-protein Kinase 33 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.29 |
Binding ≤ 10μM
|
|
TGFR2-1-E |
TGF-beta Receptor Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5600 |
0.26 |
Binding ≤ 10μM
|
|
TNI3K-1-E |
Serine/threonine-protein Kinase TNNI3K (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
55 |
0.36 |
Binding ≤ 10μM
|
|
UFO-1-E |
Tyrosine-protein Kinase Receptor UFO (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.27 |
Binding ≤ 10μM
|
|
Z104294-2-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #2 Of 2), Other |
Other |
170 |
0.34 |
Binding ≤ 10μM
|
|
Z104295-1-O |
Cyclin-dependent Kinase 4/cyclin D1 (cluster #1 Of 2), Other |
Other |
90 |
0.35 |
Binding ≤ 10μM
|
|
Z104295-1-O |
Cyclin-dependent Kinase 4/cyclin D1 (cluster #1 Of 2), Other |
Other |
100 |
0.35 |
Binding ≤ 10μM
|
|
Z104296-1-O |
Cyclin-dependent Kinase 1/cyclin B1 (cluster #1 Of 2), Other |
Other |
30 |
0.38 |
Binding ≤ 10μM
|
|
Z100090-1-O |
ABAE (Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
45 |
0.37 |
Functional ≤ 10μM
|
|
Z100091-1-O |
HS27 (Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
51 |
0.36 |
Functional ≤ 10μM
|
|
Z100093-1-O |
LX-1 (Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
75 |
0.36 |
Functional ≤ 10μM
|
|
Z100094-1-O |
MLF (Lung Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
72 |
0.36 |
Functional ≤ 10μM
|
|
Z100095-1-O |
MIP (Colon Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
120 |
0.35 |
Functional ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
86 |
0.35 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
52 |
0.36 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
46 |
0.37 |
Functional ≤ 10μM
|
|
Z80186-9-O |
K562 (Erythroleukemia Cells) (cluster #9 Of 11), Other |
Other |
130 |
0.34 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
66 |
0.36 |
Functional ≤ 10μM
|
|
Z80354-4-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #4 Of 4), Other |
Other |
54 |
0.36 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
66 |
0.36 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
77 |
0.36 |
Functional ≤ 10μM
|
|
Z80675-1-O |
AG1523 (Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
6700 |
0.26 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
96 |
0.35 |
Functional ≤ 10μM
|
|
Z80852-2-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
75 |
0.36 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
490 |
0.32 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
78 |
0.36 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
16 |
0.39 |
Functional ≤ 10μM
|
|
Z81186-1-O |
LS174T (Colon Adencocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
65 |
0.36 |
Functional ≤ 10μM
|
|
Z81225-1-O |
M109 (Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
80 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.46 |
7.04 |
-50.59 |
4 |
6 |
1 |
95 |
402.854 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.46 |
7.74 |
-91.2 |
3 |
6 |
0 |
98 |
401.846 |
2 |
↓
|
|
|
|
Analogs
-
2149675
-
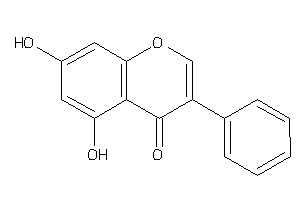
Draw
Identity
99%
90%
80%
70%
Vendors
And 50 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5000 |
0.37 |
Binding ≤ 10μM
|
|
ABCG2-1-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8800 |
0.35 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.35 |
Binding ≤ 10μM
|
|
AOFA-4-E |
Monoamine Oxidase A (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
900 |
0.42 |
Binding ≤ 10μM
|
|
AOFB-4-E |
Monoamine Oxidase B (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
900 |
0.42 |
Binding ≤ 10μM
|
|
DHB1-1-E |
Estradiol 17-beta-dehydrogenase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2210 |
0.40 |
Binding ≤ 10μM
|
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2600 |
0.39 |
Binding ≤ 10μM
|
|
ERR1-1-E |
Estrogen-related Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.56 |
Binding ≤ 10μM
|
|
ERR2-1-E |
Estrogen-related Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
395 |
0.45 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
990 |
0.42 |
Binding ≤ 10μM
|
|
ESR2-4-E |
Estrogen Receptor Beta (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
990 |
0.42 |
Binding ≤ 10μM
|
|
MGA-3-E |
Maltase-glucoamylase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2610 |
0.39 |
Binding ≤ 10μM
|
|
Q965D7-2-E |
Fatty Acid Synthase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.36 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
950 |
0.42 |
Functional ≤ 10μM
|
|
ESR2-2-E |
Estrogen Receptor Beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
956 |
0.42 |
Functional ≤ 10μM
|
|
ADO-1-E |
Aldehyde Oxidase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
340 |
0.45 |
ADME/T ≤ 10μM
|
|
Z50420-1-O |
Trypanosoma Brucei Brucei (cluster #1 Of 7), Other |
Other |
4200 |
0.38 |
Functional ≤ 10μM
|
|
Z80030-1-O |
ANN-1 (cluster #1 Of 2), Other |
Other |
8000 |
0.36 |
Functional ≤ 10μM
|
|
Z80037-1-O |
Balb/MK (cluster #1 Of 1), Other |
Other |
9100 |
0.35 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1000 |
0.42 |
Functional ≤ 10μM
|
|
Z81068-1-O |
Ishikawa (Uterine Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
510 |
0.44 |
Functional ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
956 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.27 |
1.08 |
-13.3 |
3 |
5 |
0 |
91 |
270.24 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
5.36 |
-12 |
3 |
5 |
0 |
93 |
336.391 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.59 |
Binding ≤ 10μM |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.61 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.75 |
-1.04 |
-9.47 |
3 |
4 |
0 |
70 |
179.175 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
0.75 |
-1.57 |
-12.11 |
3 |
4 |
0 |
70 |
179.175 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
400 |
0.36 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.02 |
5.63 |
-12.73 |
3 |
5 |
0 |
96 |
354.431 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.39 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
5.64 |
-12.54 |
3 |
5 |
0 |
93 |
336.391 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.38 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.43 |
3.04 |
-13.24 |
3 |
5 |
0 |
93 |
286.331 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.43 |
3.97 |
-45.94 |
2 |
5 |
-1 |
96 |
285.323 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 81 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-2-B |
Beta-lactamase (cluster #2 Of 6), Bacterial |
Bacteria |
4000 |
0.34 |
Binding ≤ 10μM
|
|
AMPH-2-B |
Penicillin-binding Protein AmpH (cluster #2 Of 2), Bacterial |
Bacteria |
4000 |
0.34 |
Binding ≤ 10μM
|
|
MDH-1-B |
Malate Dehydrogenase (cluster #1 Of 1), Bacterial |
Bacteria |
6000 |
0.33 |
Binding ≤ 10μM
|
|
NANH-1-B |
Sialidase (cluster #1 Of 1), Bacterial |
Bacteria |
9800 |
0.32 |
Binding ≤ 10μM
|
|
5NTD-1-E |
5'-nucleotidase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
45 |
0.47 |
Binding ≤ 10μM
|
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2470 |
0.36 |
Binding ≤ 10μM
|
|
AA2AR-3-E |
Adenosine A2a Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6990 |
0.33 |
Binding ≤ 10μM
|
|
ABCG2-1-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7600 |
0.33 |
Binding ≤ 10μM
|
|
AK1A1-1-E |
Aldehyde Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2320 |
0.36 |
Binding ≤ 10μM
|
|
AK1CL-1-E |
Aldo-keto Reductase Family 1 Member C21 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6900 |
0.33 |
Binding ≤ 10μM
|
|
ALDR-1-E |
Aldose Reductase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2850 |
0.35 |
Binding ≤ 10μM
|
|
AOFA-4-E |
Monoamine Oxidase A (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
10 |
0.51 |
Binding ≤ 10μM
|
|
CAH1-12-E |
Carbonic Anhydrase I (cluster #12 Of 12), Eukaryotic |
Eukaryotes |
2680 |
0.35 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
9390 |
0.32 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
9030 |
0.32 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
5410 |
0.34 |
Binding ≤ 10μM
|
|
CAH2-15-E |
Carbonic Anhydrase II (cluster #15 Of 15), Eukaryotic |
Eukaryotes |
2540 |
0.36 |
Binding ≤ 10μM
|
|
CAH3-6-E |
Carbonic Anhydrase III (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
8100 |
0.32 |
Binding ≤ 10μM
|
|
CAH4-14-E |
Carbonic Anhydrase IV (cluster #14 Of 16), Eukaryotic |
Eukaryotes |
7890 |
0.32 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
6810 |
0.33 |
Binding ≤ 10μM
|
|
CAH6-8-E |
Carbonic Anhydrase VI (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
6170 |
0.33 |
Binding ≤ 10μM
|
|
CAH7-8-E |
Carbonic Anhydrase VII (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
4840 |
0.34 |
Binding ≤ 10μM
|
|
CAH9-11-E |
Carbonic Anhydrase IX (cluster #11 Of 11), Eukaryotic |
Eukaryotes |
7000 |
0.33 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
450 |
0.40 |
Binding ≤ 10μM
|
|
CP19A-1-E |
Cytochrome P450 19A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
12 |
0.50 |
Binding ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
77 |
0.45 |
Binding ≤ 10μM
|
|
CSK21-2-E |
Casein Kinase II Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
850 |
0.39 |
Binding ≤ 10μM
|
|
CSK2B-3-E |
Casein Kinase II Beta (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
850 |
0.39 |
Binding ≤ 10μM
|
|
DHB2-1-E |
Estradiol 17-beta-dehydrogenase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1540 |
0.37 |
Binding ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
900 |
0.38 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2100 |
0.36 |
Binding ≤ 10μM
|
|
GSK3B-7-E |
Glycogen Synthase Kinase-3 Beta (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
2100 |
0.36 |
Binding ≤ 10μM
|
|
LGUL-2-E |
Glyoxalase I (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.35 |
Binding ≤ 10μM
|
|
LOX12-2-E |
Arachidonate 12-lipoxygenase (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
440 |
0.40 |
Binding ≤ 10μM
|
|
LOX15-1-E |
Arachidonate 15-lipoxygenase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2200 |
0.36 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
500 |
0.40 |
Binding ≤ 10μM
|
|
MRP1-1-E |
Multidrug Resistance-associated Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2400 |
0.36 |
Binding ≤ 10μM
|
|
NOX4-1-E |
NADPH Oxidase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
680 |
0.39 |
Binding ≤ 10μM
|
|
P85A-2-E |
PI3-kinase P85-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.34 |
Binding ≤ 10μM
|
|
P85B-2-E |
PI3-kinase P85-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.34 |
Binding ≤ 10μM
|
|
PA21B-2-E |
Phospholipase A2 Group 1B (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
PIM1-1-E |
Serine/threonine-protein Kinase PIM1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
43 |
0.47 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.34 |
Binding ≤ 10μM
|
|
PK3CB-1-E |
PI3-kinase P110-beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.34 |
Binding ≤ 10μM
|
|
PK3CD-1-E |
PI3-kinase P110-delta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.34 |
Binding ≤ 10μM
|
|
PK3CG-1-E |
PI3-kinase P110-gamma Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3800 |
0.34 |
Binding ≤ 10μM
|
|
Q965D5-1-E |
Enoyl-acyl-carrier Protein Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
473 |
0.40 |
Binding ≤ 10μM
|
|
Q965D6-1-E |
3-oxoacyl-acyl-carrier Protein Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5400 |
0.34 |
Binding ≤ 10μM
|
|
Q965D7-2-E |
Fatty Acid Synthase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Binding ≤ 10μM
|
|
TRY1-1-E |
Trypsin I (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7100 |
0.33 |
Binding ≤ 10μM
|
|
XDH-2-E |
Xanthine Dehydrogenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.38 |
Binding ≤ 10μM
|
|
LOX5-6-E |
Arachidonate 5-lipoxygenase (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
10000 |
0.32 |
Functional ≤ 10μM
|
|
CP1A1-1-E |
Cytochrome P450 1A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
660 |
0.39 |
ADME/T ≤ 10μM
|
|
CP1A2-1-E |
Cytochrome P450 1A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4097 |
0.34 |
ADME/T ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
23 |
0.49 |
ADME/T ≤ 10μM
|
|
Z102178-2-O |
Liver Microsomes (cluster #2 Of 2), Other |
Other |
7500 |
0.33 |
Functional ≤ 10μM
|
|
Z102342-1-O |
Liver (cluster #1 Of 1), Other |
Other |
6000 |
0.33 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
8900 |
0.32 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
8500 |
0.32 |
Functional ≤ 10μM
|
|
Z80418-2-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #2 Of 9), Other |
Other |
9600 |
0.32 |
Functional ≤ 10μM
|
|
Z81000-1-O |
HT-22 (Hippocampal Cells) (cluster #1 Of 1), Other |
Other |
2980 |
0.35 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
5000 |
0.34 |
Functional ≤ 10μM
|
|
R1AB-1-V |
Replicase Polyprotein 1ab (cluster #1 Of 1), Viral |
Viruses |
8100 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.68 |
-2.9 |
-13.58 |
5 |
7 |
0 |
131 |
302.238 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.94 |
-2.61 |
-43.8 |
4 |
7 |
-1 |
134 |
301.23 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
700 |
0.36 |
Binding ≤ 10μM |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.33 |
Functional ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.36 |
Functional ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
660 |
0.36 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
7943 |
0.30 |
Functional ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
2500 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
4.57 |
-11.8 |
3 |
5 |
0 |
93 |
322.364 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1500 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
7.57 |
-10.07 |
0 |
4 |
0 |
39 |
325.408 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1500 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
10.36 |
-44.34 |
1 |
3 |
1 |
31 |
284.379 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
860 |
0.37 |
Binding ≤ 10μM
|
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.33 |
Functional ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
3500 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
3.86 |
-10.95 |
3 |
5 |
0 |
93 |
308.337 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
3.88 |
-10.98 |
3 |
5 |
0 |
93 |
308.337 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2100 |
0.35 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4900 |
0.32 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
4.63 |
-12.46 |
2 |
5 |
0 |
85 |
306.321 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
3.02 |
-11.13 |
3 |
5 |
0 |
93 |
294.31 |
4 |
↓
|
|
|
|
Analogs
-
12352660
-
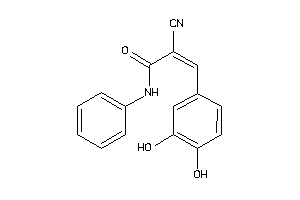
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
700 |
0.41 |
Binding ≤ 10μM |
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.22 |
2.9 |
-11.76 |
3 |
5 |
0 |
93 |
280.283 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7460 |
0.45 |
Binding ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
2420 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
4.73 |
-18.03 |
1 |
4 |
0 |
54 |
298.165 |
3 |
↓
|
|
|
|
|
|
|
Analogs
-
17045393
-
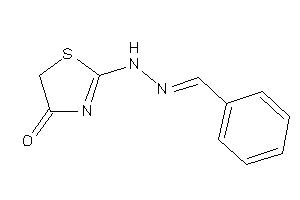
-
13633783
-
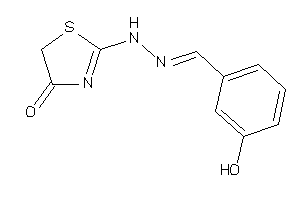
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4710 |
0.47 |
Binding ≤ 10μM
|
|
ERBB2-3-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6930 |
0.45 |
Binding ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1670 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.21 |
1.27 |
-17.58 |
2 |
5 |
0 |
74 |
235.268 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
0.67 |
2.25 |
-10.41 |
2 |
5 |
0 |
74 |
235.268 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.67 |
3 |
-47.3 |
1 |
5 |
-1 |
77 |
234.26 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1720 |
0.50 |
Binding ≤ 10μM
|
|
ERBB2-3-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2430 |
0.49 |
Binding ≤ 10μM
|
|
Z80224-12-O |
MCF7 (Breast Carcinoma Cells) (cluster #12 Of 14), Other |
Other |
970 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.49 |
4.74 |
-15.97 |
1 |
4 |
0 |
54 |
298.165 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
1.96 |
5.69 |
-7.92 |
1 |
4 |
0 |
54 |
298.165 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
90 |
0.58 |
Binding ≤ 10μM
|
|
ERBB2-3-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
420 |
0.53 |
Binding ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
60 |
0.59 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
3.19 |
-8.36 |
2 |
5 |
0 |
74 |
314.164 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
1.88 |
3.23 |
-9.1 |
2 |
5 |
0 |
74 |
314.164 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
2.41 |
2.24 |
-14.82 |
2 |
5 |
0 |
74 |
314.164 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1460 |
0.45 |
Binding ≤ 10μM
|
|
ERBB2-3-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4510 |
0.42 |
Binding ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1560 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.68 |
2.68 |
-16.14 |
2 |
5 |
0 |
74 |
304.158 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.14 |
4.43 |
-41.6 |
1 |
5 |
-1 |
77 |
303.15 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.68 |
3.46 |
-50.77 |
1 |
5 |
-1 |
77 |
303.15 |
3 |
↓
|
|
|
|
|
|
|
Analogs
-
5124238
-
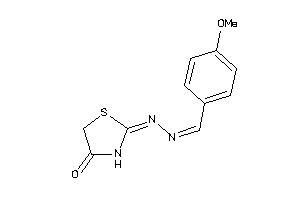
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5780 |
0.43 |
Binding ≤ 10μM
|
|
ERBB2-3-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
9170 |
0.41 |
Binding ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
3160 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.21 |
4.4 |
-10.31 |
1 |
5 |
0 |
63 |
249.295 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
1.74 |
3.43 |
-17.55 |
1 |
5 |
0 |
63 |
249.295 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
1.21 |
4.37 |
-9.47 |
1 |
5 |
0 |
63 |
249.295 |
3 |
↓
|
|
|
|
Analogs
-
8582462
-
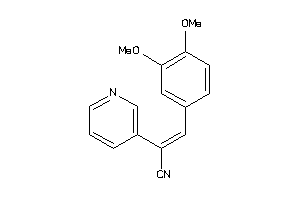
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
700 |
0.43 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
7.01 |
-11.36 |
0 |
4 |
0 |
55 |
266.3 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
850 |
0.57 |
Binding ≤ 10μM
|
|
ERBB2-1-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5700 |
0.49 |
Binding ≤ 10μM
|
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.33 |
0.14 |
-13.55 |
4 |
4 |
0 |
90 |
220.253 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
9400 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.96 |
2.07 |
-9.56 |
2 |
5 |
0 |
86 |
195.174 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.54 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.14 |
1.65 |
-10.2 |
2 |
5 |
0 |
86 |
181.147 |
2 |
↓
|
|
|
|
Analogs
-
3199665
-
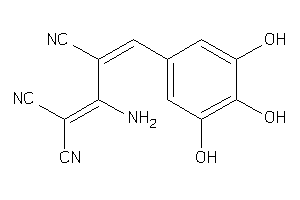
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
800 |
0.43 |
Binding ≤ 10μM
|
|
FGFR1-1-E |
Fibroblast Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.37 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2800 |
0.39 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4300 |
0.38 |
Binding ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
6310 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.67 |
-1.34 |
-28.09 |
5 |
7 |
0 |
158 |
268.232 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.67 |
-0.43 |
-54.85 |
4 |
7 |
-1 |
161 |
267.224 |
2 |
↓
|
|
|
|
Analogs
-
160515
-
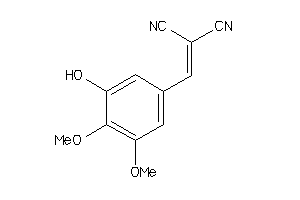
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.46 |
Binding ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
10000 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.22 |
1.25 |
-14.76 |
2 |
5 |
0 |
97 |
216.196 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.22 |
2.18 |
-54.37 |
1 |
5 |
-1 |
100 |
215.188 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.22 |
2.28 |
-51.15 |
1 |
5 |
-1 |
100 |
215.188 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6500 |
0.45 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6500 |
0.45 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6500 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.09 |
2.65 |
-7.78 |
3 |
4 |
0 |
83 |
211.224 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.40 |
1.25 |
-44.94 |
2 |
4 |
-1 |
86 |
210.216 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6500 |
0.45 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6500 |
0.45 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6500 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
5.11 |
-9.73 |
3 |
3 |
0 |
66 |
227.292 |
2 |
↓
|
|
|
|
Analogs
-
4151288
-
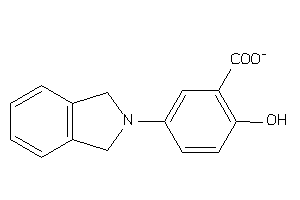
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4100 |
0.27 |
Binding ≤ 10μM
|
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.26 |
Functional ≤ 10μM
|
|
ERBB2-1-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9000 |
0.25 |
Functional ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.12 |
3.35 |
-54.38 |
4 |
7 |
-1 |
124 |
380.376 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3300 |
0.38 |
Binding ≤ 10μM
|
|
ERBB2-1-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
Functional ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
-3.72 |
-58.16 |
3 |
5 |
-1 |
100 |
273.264 |
4 |
↓
|
|
|
|
Analogs
-
39119523
-
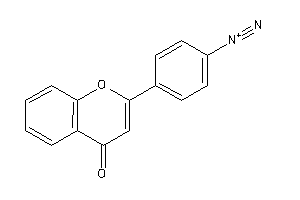
-
39119546
-
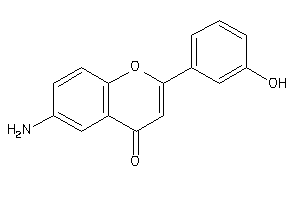
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8700 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.87 |
4.03 |
-12.31 |
4 |
4 |
0 |
82 |
252.273 |
1 |
↓
|
|