|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
16 |
1.36 |
Binding ≤ 10μM
|
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
1.58 |
Binding ≤ 10μM |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
1.17 |
Functional ≤ 10μM |
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
1.17 |
Functional ≤ 10μM |
|
Z50425-5-O |
Plasmodium Falciparum (cluster #5 Of 22), Other |
Other |
3162 |
0.96 |
Functional ≤ 10μM
|
|
Z50597-2-O |
Rattus Norvegicus (cluster #2 Of 12), Other |
Other |
450 |
1.11 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.55 |
-0.77 |
-49.63 |
3 |
3 |
0 |
68 |
137.119 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
16 |
1.36 |
Binding ≤ 10μM
|
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
1.58 |
Binding ≤ 10μM |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
1.17 |
Functional ≤ 10μM |
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
1.17 |
Functional ≤ 10μM |
|
Z50425-5-O |
Plasmodium Falciparum (cluster #5 Of 22), Other |
Other |
3162 |
0.96 |
Functional ≤ 10μM
|
|
Z50597-2-O |
Rattus Norvegicus (cluster #2 Of 12), Other |
Other |
450 |
1.11 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
|
|
|
|
|
|
Analogs
-
85733
-
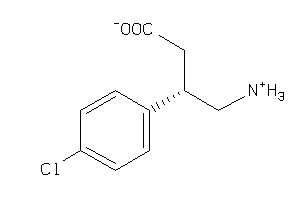
Draw
Identity
99%
90%
80%
70%
Vendors
And 64 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R-3-E |
Adenosine Receptor A3 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
50 |
0.73 |
Binding ≤ 10μM
|
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
32 |
0.75 |
Binding ≤ 10μM
|
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
60 |
0.72 |
Binding ≤ 10μM
|
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
32 |
0.75 |
Binding ≤ 10μM
|
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
60 |
0.72 |
Binding ≤ 10μM
|
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
330 |
0.65 |
Binding ≤ 10μM
|
|
GBRB2-4-E |
GABA Receptor Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
330 |
0.65 |
Binding ≤ 10μM
|
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
330 |
0.65 |
Binding ≤ 10μM
|
|
Z104301-5-O |
GABA-A Receptor; Anion Channel (cluster #5 Of 8), Other |
Other |
30 |
0.75 |
Binding ≤ 10μM
|
|
Z50512-6-O |
Cavia Porcellus (cluster #6 Of 7), Other |
Other |
6400 |
0.52 |
Functional ≤ 10μM |
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
330 |
0.65 |
Functional ≤ 10μM
|
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
1800 |
0.57 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.42 |
4.81 |
-48.33 |
3 |
3 |
0 |
68 |
213.664 |
4 |
↓
|
|
|
|
Analogs
-
61
-
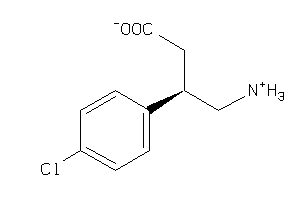
Draw
Identity
99%
90%
80%
70%
Vendors
And 53 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.42 |
4.85 |
-63 |
3 |
3 |
0 |
68 |
213.664 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.74 |
3.5 |
-48.45 |
3 |
3 |
0 |
68 |
219.265 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1050 |
0.93 |
Binding ≤ 10μM
|
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1050 |
0.93 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.20 |
-4.12 |
-57.39 |
4 |
4 |
0 |
88 |
153.118 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1050 |
0.93 |
Binding ≤ 10μM
|
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1050 |
0.93 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.20 |
-4.12 |
-57.38 |
4 |
4 |
0 |
88 |
153.118 |
4 |
↓
|
|
|
|
Analogs
-
13742937
-
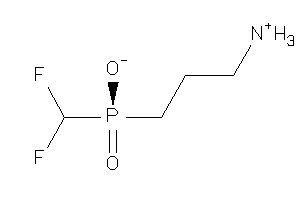
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
89 |
0.99 |
Binding ≤ 10μM
|
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
89 |
0.99 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.00 |
-0.4 |
-52.87 |
3 |
3 |
0 |
68 |
173.099 |
4 |
↓
|
|
|
|
Analogs
-
13742936
-
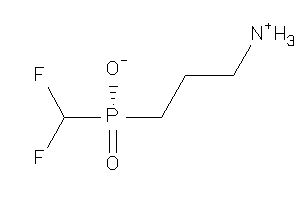
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
89 |
0.99 |
Binding ≤ 10μM
|
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
89 |
0.99 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.00 |
-0.4 |
-52.84 |
3 |
3 |
0 |
68 |
173.099 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5600 |
0.41 |
Binding ≤ 10μM
|
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5600 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.97 |
-1.58 |
-53.03 |
3 |
5 |
0 |
90 |
249.266 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5600 |
0.41 |
Binding ≤ 10μM
|
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5600 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.97 |
-1.58 |
-68.47 |
3 |
5 |
0 |
90 |
249.266 |
5 |
↓
|
|
|
|
Analogs
-
1720869
-
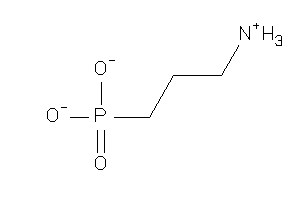
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6810 |
0.90 |
Binding ≤ 10μM
|
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6810 |
0.90 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.87 |
-2.45 |
-50.53 |
4 |
4 |
0 |
88 |
139.091 |
3 |
↓
|
|
|
|
Analogs
-
6093247
-
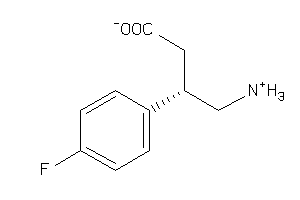
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1330 |
0.59 |
Binding ≤ 10μM
|
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1330 |
0.59 |
Binding ≤ 10μM
|
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
800 |
0.61 |
Binding ≤ 10μM
|
|
GBRB2-4-E |
GABA Receptor Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
800 |
0.61 |
Binding ≤ 10μM
|
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
800 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.93 |
-1.06 |
-53.62 |
3 |
3 |
0 |
67 |
197.209 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.25 |
3.98 |
-51.06 |
3 |
3 |
0 |
68 |
227.244 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2500 |
0.56 |
Binding ≤ 10μM |
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2500 |
0.56 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2500 |
0.56 |
Binding ≤ 10μM |
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2500 |
0.56 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.87 |
0.51 |
-51.57 |
3 |
5 |
0 |
86 |
225.225 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.61 |
Binding ≤ 10μM
|
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.94 |
3.82 |
-51.71 |
3 |
3 |
0 |
68 |
213.664 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.61 |
Binding ≤ 10μM
|
|
GABR2-1-E |
GABA-B Receptor 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.61 |
Binding ≤ 10μM
|
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
800 |
0.61 |
Binding ≤ 10μM
|
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
800 |
0.61 |
Binding ≤ 10μM
|
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
800 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.94 |
3.81 |
-51.07 |
3 |
3 |
0 |
68 |
213.664 |
4 |
↓
|
|