|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
21 |
0.51 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
550 |
0.42 |
Binding ≤ 10μM
|
|
EPHB4-1-E |
Ephrin Type-B Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.44 |
Binding ≤ 10μM
|
|
HCK-1-E |
Tyrosine-protein Kinase HCK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.56 |
Binding ≤ 10μM
|
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.48 |
Binding ≤ 10μM
|
|
PI4KB-1-E |
PI4-kinase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
17 |
0.52 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
46 |
0.49 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
240 |
0.44 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
27 |
0.50 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM
|
|
PRKDC-1-E |
DNA-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.52 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.53 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
55 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
5.36 |
-8.75 |
3 |
6 |
0 |
90 |
348.204 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.62 |
6.12 |
-43.42 |
2 |
6 |
-1 |
93 |
347.196 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
410 |
0.39 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4400 |
0.33 |
Binding ≤ 10μM
|
|
EPHB4-1-E |
Ephrin Type-B Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.33 |
Binding ≤ 10μM
|
|
HCK-1-E |
Tyrosine-protein Kinase HCK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.36 |
Binding ≤ 10μM
|
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.49 |
Binding ≤ 10μM |
|
PI4KB-1-E |
PI4-kinase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
408 |
0.39 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.35 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.34 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.43 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1300 |
0.36 |
Binding ≤ 10μM
|
|
PRKDC-1-E |
DNA-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
410 |
0.39 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
5.11 |
-9.67 |
4 |
7 |
0 |
106 |
308.345 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
2.19 |
6.1 |
-14.78 |
6 |
7 |
0 |
108 |
310.361 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.19 |
5.6 |
-10.5 |
5 |
7 |
0 |
107 |
309.353 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3200 |
0.28 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
579 |
0.32 |
Binding ≤ 10μM
|
|
EPHB4-1-E |
Ephrin Type-B Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2910 |
0.29 |
Binding ≤ 10μM
|
|
HCK-1-E |
Tyrosine-protein Kinase HCK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2400 |
0.29 |
Binding ≤ 10μM
|
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
80 |
0.37 |
Binding ≤ 10μM
|
|
PI4KB-1-E |
PI4-kinase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5700 |
0.27 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.29 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5800 |
0.27 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
680 |
0.32 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
990 |
0.31 |
Binding ≤ 10μM
|
|
PRKDC-1-E |
DNA-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
339 |
0.34 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3200 |
0.28 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1680 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
7.35 |
-20.22 |
3 |
8 |
0 |
111 |
381.465 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MRCKA-1-E |
Serine/threonine-protein Kinase MRCK-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.16 |
Binding ≤ 10μM
|
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.27 |
Binding ≤ 10μM
|
|
P3C2A-1-E |
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Subunit Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
176 |
0.21 |
Binding ≤ 10μM
|
|
P3C2B-1-E |
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Beta Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
549 |
0.19 |
Binding ≤ 10μM
|
|
P85A-2-E |
PI3-kinase P85-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
564 |
0.19 |
Binding ≤ 10μM
|
|
PI4KB-1-E |
PI4-kinase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6680 |
0.16 |
Binding ≤ 10μM
|
|
PK3C3-1-E |
Phosphatidylinositol 3-kinase Catalytic Subunit Type 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
533 |
0.20 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
250 |
0.21 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
564 |
0.19 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
171 |
0.21 |
Binding ≤ 10μM
|
|
PRKDC-1-E |
DNA-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.83 |
15.43 |
-21.37 |
0 |
7 |
0 |
71 |
607.636 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.41 |
4.79 |
-7.76 |
2 |
8 |
0 |
109 |
326.36 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.37 |
Binding ≤ 10μM |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.39 |
Binding ≤ 10μM |
|
PK3C3-1-E |
Phosphatidylinositol 3-kinase Catalytic Subunit Type 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
31 |
0.34 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM |
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.39 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Binding ≤ 10μM
|
|
PRKDC-1-E |
DNA-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.38 |
Binding ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
47 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.29 |
6.65 |
-50.42 |
1 |
7 |
-1 |
103 |
475.934 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.29 |
6.66 |
-19.89 |
2 |
7 |
0 |
101 |
476.942 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.27 |
Binding ≤ 10μM |
|
PI4KB-1-E |
PI4-kinase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM |
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.35 |
Binding ≤ 10μM |
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
92 |
0.35 |
Binding ≤ 10μM |
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.38 |
Binding ≤ 10μM |
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM |
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
730 |
0.31 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
6.37 |
-11.32 |
3 |
10 |
0 |
124 |
380.412 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.18 |
6.63 |
-36.84 |
4 |
10 |
1 |
125 |
381.42 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.30 |
5.16 |
-16.72 |
2 |
10 |
0 |
127 |
377.43 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Binding ≤ 10μM
|
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.38 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
5.88 |
-54.66 |
1 |
8 |
-1 |
112 |
487.97 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.18 |
5.88 |
-20.98 |
2 |
8 |
0 |
110 |
488.978 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5300 |
0.35 |
Binding ≤ 10μM
|
|
PRKDC-1-E |
DNA-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
280 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.26 |
6.84 |
-11.8 |
0 |
5 |
0 |
47 |
281.315 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.37 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.39 |
9.2 |
-16.08 |
1 |
9 |
0 |
93 |
465.554 |
5 |
↓
|
|
|
|
Analogs
-
14975141
-
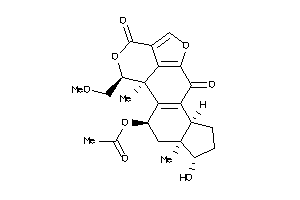
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
193 |
0.30 |
Binding ≤ 10μM
|
|
P85B-2-E |
PI3-kinase P85-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM |
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM |
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.38 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM |
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM |
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
PK3CG-3-E |
PI3-kinase P110-gamma Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM |
|
PK3CG-3-E |
PI3-kinase P110-gamma Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
10 |
0.36 |
Functional ≤ 10μM |
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
1464 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
1.2 |
-13.15 |
1 |
8 |
0 |
112 |
430.453 |
4 |
↓
|
|
|
|
Analogs
-
3995884
-
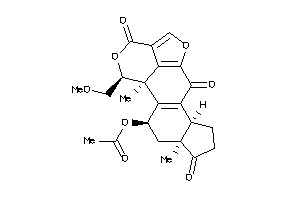
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.33 |
Binding ≤ 10μM
|
|
MYLK-1-E |
Myosin Light Chain Kinase, Smooth Muscle (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.24 |
Binding ≤ 10μM
|
|
P85A-2-E |
PI3-kinase P85-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
P85B-2-E |
PI3-kinase P85-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.36 |
Binding ≤ 10μM
|
|
PK3CG-3-E |
PI3-kinase P110-gamma Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
89 |
0.32 |
Binding ≤ 10μM
|
|
PLK1-1-E |
Serine/threonine-protein Kinase PLK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.37 |
Binding ≤ 10μM
|
|
PLK3-1-E |
Serine/threonine-protein Kinase PLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
49 |
0.33 |
Binding ≤ 10μM
|
|
PRKDC-1-E |
DNA-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.31 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.23 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
98 |
0.32 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.84 |
8.68 |
-13.81 |
0 |
8 |
0 |
109 |
428.437 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3400 |
0.35 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3400 |
0.35 |
Binding ≤ 10μM
|
|
KAPCG-1-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.35 |
Binding ≤ 10μM
|
|
KCC2A-1-E |
CaM Kinase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
370 |
0.41 |
Binding ≤ 10μM
|
|
KCC2B-1-E |
CaM Kinase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
370 |
0.41 |
Binding ≤ 10μM
|
|
KCC2G-1-E |
CaM Kinase II Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
370 |
0.41 |
Binding ≤ 10μM
|
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
49 |
0.47 |
Binding ≤ 10μM
|
|
P3C2B-1-E |
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Beta Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.42 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.54 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
16 |
0.50 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
660 |
0.39 |
Binding ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
580 |
0.40 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
270 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
1300 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
4.03 |
-14.97 |
1 |
5 |
0 |
58 |
313.382 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.98 |
4.35 |
-28.48 |
2 |
5 |
1 |
60 |
314.39 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.29 |
Binding ≤ 10μM
|
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
570 |
0.25 |
Binding ≤ 10μM
|
|
NTRK1-1-E |
Nerve Growth Factor Receptor Trk-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2850 |
0.22 |
Binding ≤ 10μM
|
|
P3C2B-1-E |
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Beta Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
670 |
0.25 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
33 |
0.30 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.28 |
Binding ≤ 10μM
|
|
PRKDC-1-E |
DNA-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1230 |
0.24 |
Binding ≤ 10μM
|
|
Z80240-1-O |
MDA-MB-361 (cluster #1 Of 1), Other |
Other |
720 |
0.25 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
390 |
0.26 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
140 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
5.07 |
-19.39 |
1 |
10 |
0 |
108 |
513.649 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.44 |
7.29 |
-60.13 |
2 |
10 |
1 |
109 |
514.657 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.30 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
801 |
0.22 |
Binding ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
213 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
10.86 |
-50.55 |
2 |
11 |
1 |
112 |
529.625 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.32 |
Binding ≤ 10μM |
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1026 |
0.23 |
Binding ≤ 10μM |
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6315 |
0.20 |
Binding ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
355 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.25 |
7.96 |
-15.65 |
1 |
12 |
0 |
124 |
495.54 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.28 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.21 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
945 |
0.19 |
Binding ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
1 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.78 |
13.77 |
-50.7 |
3 |
11 |
1 |
105 |
622.676 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.78 |
11.41 |
-16.49 |
2 |
11 |
0 |
104 |
621.668 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1962 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
12.68 |
-50.19 |
2 |
9 |
1 |
89 |
495.611 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.29 |
Binding ≤ 10μM |
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.30 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.27 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
16 |
0.26 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.27 |
Binding ≤ 10μM
|
|
Z80240-1-O |
MDA-MB-361 (cluster #1 Of 1), Other |
Other |
8 |
0.27 |
Functional ≤ 10μM |
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
21 |
0.26 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.38 |
10.66 |
-59.19 |
3 |
13 |
1 |
135 |
571.666 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.38 |
8.31 |
-22.29 |
2 |
13 |
0 |
134 |
570.658 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.33 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.31 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
49 |
0.28 |
Binding ≤ 10μM
|
|
Z80240-1-O |
MDA-MB-361 (cluster #1 Of 1), Other |
Other |
816 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.15 |
13.22 |
-61.79 |
2 |
12 |
-1 |
145 |
504.527 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.15 |
13.5 |
-73.89 |
3 |
12 |
0 |
146 |
505.535 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
13 |
0.29 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
85 |
0.26 |
Binding ≤ 10μM
|
|
Z80240-1-O |
MDA-MB-361 (cluster #1 Of 1), Other |
Other |
115 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.41 |
13.33 |
-18.25 |
2 |
12 |
0 |
131 |
519.562 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.41 |
13.58 |
-38.77 |
3 |
12 |
1 |
132 |
520.57 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1180 |
0.22 |
Binding ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
2 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
8.9 |
-15.57 |
2 |
11 |
0 |
116 |
519.606 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
45 |
0.34 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
16 |
0.36 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
44 |
0.34 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.39 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
49 |
0.34 |
Binding ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
210 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.14 |
-4.94 |
-11.62 |
0 |
9 |
0 |
81 |
417.42 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
P3C2A-1-E |
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Subunit Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
28 |
0.33 |
Binding ≤ 10μM
|
|
P3C2B-1-E |
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Beta Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.33 |
Binding ≤ 10μM
|
|
P85A-2-E |
PI3-kinase P85-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Binding ≤ 10μM
|
|
PI4KB-1-E |
PI4-kinase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
18 |
0.34 |
Binding ≤ 10μM
|
|
PK3C3-1-E |
Phosphatidylinositol 3-kinase Catalytic Subunit Type 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.35 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
18 |
0.34 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.36 |
Binding ≤ 10μM
|
|
PRKDC-1-E |
DNA-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.29 |
9.49 |
-15.63 |
2 |
5 |
0 |
74 |
432.405 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.29 |
10.02 |
-48.2 |
3 |
5 |
1 |
75 |
433.413 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.38 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
250 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.41 |
9.56 |
-15.45 |
2 |
5 |
0 |
74 |
450.395 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.41 |
9.89 |
-44.58 |
3 |
5 |
1 |
75 |
451.403 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.35 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8900 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.46 |
9.22 |
-17.45 |
1 |
9 |
0 |
93 |
465.554 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.51 |
Binding ≤ 10μM
|
|
BCR-1-E |
Breakpoint Cluster Region Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
18 |
0.45 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
260 |
0.38 |
Binding ≤ 10μM
|
|
EPHB4-1-E |
Ephrin Type-B Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
190 |
0.39 |
Binding ≤ 10μM
|
|
HCK-1-E |
Tyrosine-protein Kinase HCK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.47 |
Binding ≤ 10μM
|
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.47 |
Binding ≤ 10μM
|
|
PI4KB-1-E |
PI4-kinase Beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.42 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
52 |
0.42 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.34 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.40 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.35 |
Binding ≤ 10μM
|
|
PRKDC-1-E |
DNA-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
60 |
0.42 |
Binding ≤ 10μM
|
|
RET-1-E |
Tyrosine-protein Kinase Receptor RET (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
14 |
0.46 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.43 |
Binding ≤ 10μM
|
|
Z81057-4-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #4 Of 4), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.24 |
8.08 |
-10.14 |
3 |
7 |
0 |
98 |
319.372 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.24 |
8.38 |
-34.95 |
4 |
7 |
1 |
100 |
320.38 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.24 |
4 |
-34.3 |
4 |
7 |
1 |
100 |
320.38 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KSYK-1-E |
Tyrosine-protein Kinase SYK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
371 |
0.35 |
Binding ≤ 10μM
|
|
M3K9-1-E |
Mitogen-activated Protein Kinase Kinase Kinase 9 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
591 |
0.34 |
Binding ≤ 10μM
|
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.41 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.43 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
16 |
0.42 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.42 |
Binding ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
330 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.90 |
4.2 |
-10.3 |
3 |
8 |
0 |
110 |
372.454 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.90 |
4.55 |
-27.71 |
4 |
8 |
1 |
112 |
373.462 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
750 |
0.32 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.44 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.39 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.43 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
15 |
0.41 |
Binding ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.12 |
4.54 |
-10.38 |
3 |
8 |
0 |
110 |
386.481 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.12 |
5.53 |
-26.57 |
4 |
8 |
1 |
112 |
387.489 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
21 |
0.32 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.35 |
Binding ≤ 10μM
|
|
PK3CB-2-E |
PI3-kinase P110-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.29 |
Binding ≤ 10μM
|
|
PK3CD-2-E |
PI3-kinase P110-delta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.34 |
Binding ≤ 10μM
|
|
PK3CG-2-E |
PI3-kinase P110-gamma Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
15 |
0.32 |
Binding ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
174 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.20 |
3.9 |
-17 |
2 |
11 |
0 |
131 |
504.642 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.20 |
6.11 |
-58.96 |
3 |
11 |
1 |
132 |
505.65 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.20 |
4.24 |
-35.5 |
3 |
11 |
1 |
132 |
505.65 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.94 |
14.07 |
-51.7 |
3 |
11 |
1 |
109 |
625.676 |
10 |
↓
|
|