|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6500 |
0.24 |
Binding ≤ 10μM
|
|
RARB-1-E |
Retinoic Acid Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2480 |
0.26 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
77 |
0.33 |
Binding ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
280 |
0.31 |
Functional ≤ 10μM
|
|
Z80135-1-O |
F9 (cluster #1 Of 1), Other |
Other |
33 |
0.35 |
Functional ≤ 10μM
|
|
Z80140-1-O |
GBM (cluster #1 Of 1), Other |
Other |
550 |
0.29 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
490 |
0.29 |
Functional ≤ 10μM
|
|
Z80250-1-O |
Me665/2/21 (cluster #1 Of 1), Other |
Other |
420 |
0.30 |
Functional ≤ 10μM
|
|
Z80306-1-O |
NB-4 (Promyelocytic Leukemia Cells) (cluster #1 Of 1), Other |
Other |
400 |
0.30 |
Functional ≤ 10μM
|
|
Z80354-1-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
130 |
0.32 |
Functional ≤ 10μM |
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
430 |
0.30 |
Functional ≤ 10μM
|
|
Z80449-1-O |
SAOS-2 (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
670 |
0.29 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
440 |
0.30 |
Functional ≤ 10μM
|
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
510 |
0.29 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
680 |
0.29 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
430 |
0.30 |
Functional ≤ 10μM
|
|
Z81065-1-O |
IGROV-1 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
350 |
0.30 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
320 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.63 |
13.65 |
-51.5 |
1 |
3 |
-1 |
60 |
397.494 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 53 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
832 |
0.34 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Binding ≤ 10μM
|
|
PPARD-2-E |
Peroxisome Proliferator-activated Receptor Delta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3620 |
0.30 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.44 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.40 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
325 |
0.36 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Functional ≤ 10μM
|
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
76 |
0.40 |
Functional ≤ 10μM
|
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
210 |
0.37 |
Functional ≤ 10μM
|
|
Z80106-1-O |
COS-1 (Kidney Cells) (cluster #1 Of 1), Other |
Other |
23 |
0.43 |
Functional ≤ 10μM
|
|
Z80169-1-O |
Huh-7 (Hepatocellular Carcinoma) (cluster #1 Of 1), Other |
Other |
220 |
0.37 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
30 |
0.42 |
Functional ≤ 10μM
|
|
Z81117-1-O |
Keratinocytes (Keratinocytes) (cluster #1 Of 2), Other |
Other |
8000 |
0.29 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
5000 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
7.4 |
-33.54 |
2 |
6 |
1 |
73 |
358.443 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.35 |
7.28 |
-11.07 |
1 |
6 |
0 |
72 |
357.435 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 54 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
832 |
0.34 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Binding ≤ 10μM
|
|
PPARD-2-E |
Peroxisome Proliferator-activated Receptor Delta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3620 |
0.30 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.44 |
Binding ≤ 10μM
|
|
PPARG-1-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.41 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
325 |
0.36 |
Binding ≤ 10μM
|
|
PPARA-1-E |
Peroxisome Proliferator-activated Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Functional ≤ 10μM
|
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
76 |
0.40 |
Functional ≤ 10μM
|
|
Z80106-1-O |
COS-1 (Kidney Cells) (cluster #1 Of 1), Other |
Other |
23 |
0.43 |
Functional ≤ 10μM
|
|
Z80169-1-O |
Huh-7 (Hepatocellular Carcinoma) (cluster #1 Of 1), Other |
Other |
220 |
0.37 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
30 |
0.42 |
Functional ≤ 10μM
|
|
Z81117-1-O |
Keratinocytes (Keratinocytes) (cluster #1 Of 2), Other |
Other |
8000 |
0.29 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
5000 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
7.4 |
-33.23 |
2 |
6 |
1 |
73 |
358.443 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.35 |
7.28 |
-11.54 |
1 |
6 |
0 |
72 |
357.435 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CDC7-1-E |
Cell Division Cycle 7-related Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.70 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
250 |
0.58 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
240 |
0.58 |
Binding ≤ 10μM
|
|
CDK5-1-E |
Cyclin-dependent Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
460 |
0.55 |
Binding ≤ 10μM
|
|
CDK9-1-E |
Cyclin-dependent Kinase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
34 |
0.65 |
Binding ≤ 10μM
|
|
CHK2-1-E |
Serine/threonine-protein Kinase Chk2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.52 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
220 |
0.58 |
Binding ≤ 10μM
|
|
KS6A4-1-E |
Ribosomal Protein S6 Kinase Alpha 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3010 |
0.48 |
Binding ≤ 10μM
|
|
KS6A5-2-E |
Ribosomal Protein S6 Kinase Alpha 5 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2340 |
0.49 |
Binding ≤ 10μM
|
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.55 |
Binding ≤ 10μM
|
|
MAPK5-1-E |
MAP Kinase-activated Protein Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Binding ≤ 10μM
|
|
MKNK1-1-E |
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2670 |
0.49 |
Binding ≤ 10μM
|
|
MKNK2-1-E |
MAP Kinase Signal-integrating Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
171 |
0.59 |
Binding ≤ 10μM
|
|
PLK1-1-E |
Serine/threonine-protein Kinase PLK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
980 |
0.53 |
Binding ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
5000 |
0.46 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
5870 |
0.46 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
1330 |
0.51 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
4590 |
0.47 |
Functional ≤ 10μM
|
|
Z80466-1-O |
SF268 (cluster #1 Of 4), Other |
Other |
860 |
0.53 |
Functional ≤ 10μM
|
|
Z80525-1-O |
SW48 (cluster #1 Of 2), Other |
Other |
1200 |
0.52 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
2670 |
0.49 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
1490 |
0.51 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
970 |
0.53 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
1100 |
0.52 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
3200 |
0.48 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
1830 |
0.50 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
3810 |
0.47 |
Functional ≤ 10μM
|
|
Z80335-1-O |
NHDF (cluster #1 Of 2), Other |
Other |
1600 |
0.51 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.64 |
2.58 |
-15.17 |
2 |
4 |
0 |
58 |
213.24 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.64 |
2.88 |
-44.62 |
3 |
4 |
1 |
59 |
214.248 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.96 |
13.74 |
-19.57 |
0 |
8 |
0 |
77 |
547.055 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.96 |
15.44 |
-53.42 |
1 |
8 |
1 |
78 |
548.063 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.96 |
13.26 |
-20.42 |
0 |
8 |
0 |
77 |
547.055 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.96 |
15.49 |
-56.83 |
1 |
8 |
1 |
78 |
548.063 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
D3ZWV0-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
Q499N1-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.33 |
Binding ≤ 10μM
|
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.33 |
Binding ≤ 10μM
|
|
RARB-1-E |
Retinoic Acid Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.33 |
Binding ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
100 |
0.35 |
Functional ≤ 10μM
|
|
Z80140-1-O |
GBM (cluster #1 Of 1), Other |
Other |
180 |
0.34 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
150 |
0.34 |
Functional ≤ 10μM
|
|
Z80250-1-O |
Me665/2/21 (cluster #1 Of 1), Other |
Other |
250 |
0.33 |
Functional ≤ 10μM
|
|
Z80306-1-O |
NB-4 (Promyelocytic Leukemia Cells) (cluster #1 Of 1), Other |
Other |
82 |
0.35 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
210 |
0.33 |
Functional ≤ 10μM
|
|
Z80449-1-O |
SAOS-2 (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
250 |
0.33 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
260 |
0.33 |
Functional ≤ 10μM
|
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
250 |
0.33 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
320 |
0.32 |
Functional ≤ 10μM
|
|
Z81024-5-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #5 Of 8), Other |
Other |
190 |
0.34 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
100 |
0.35 |
Functional ≤ 10μM
|
|
Z81065-3-O |
IGROV-1 (Ovarian Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
230 |
0.33 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
120 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.53 |
0.19 |
-51.44 |
1 |
3 |
-1 |
60 |
373.472 |
4 |
↓
|
|
|
|
Analogs
-
1530090
-
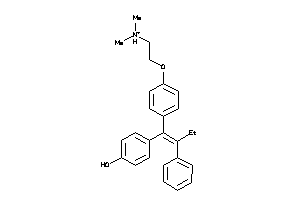
-
1530689
-
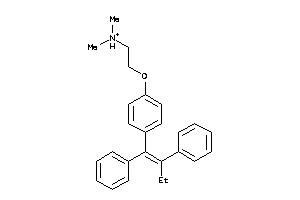
-
1530690
-
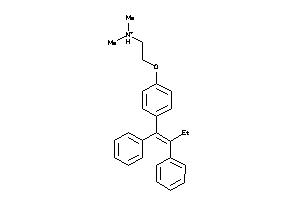
-
8602413
-
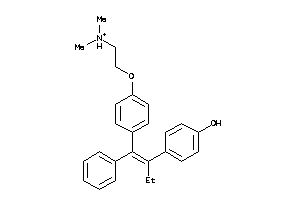
-
44699472
-
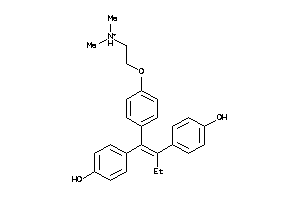
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ERR3-1-E |
Estrogen-related Receptor Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
13 |
0.38 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6 |
0.40 |
Binding ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
32 |
0.36 |
Binding ≤ 10μM
|
|
PLD1-1-E |
Phospholipase D1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4800 |
0.26 |
Binding ≤ 10μM
|
|
PLD2-1-E |
Phospholipase D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
44 |
0.36 |
Binding ≤ 10μM
|
|
ERR3-1-E |
Estrogen-related Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.37 |
Functional ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.39 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.39 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
8000 |
0.25 |
Functional ≤ 10μM
|
|
Z80296-1-O |
MVLN (cluster #1 Of 1), Other |
Other |
3 |
0.41 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
2 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.59 |
12.96 |
-37.36 |
2 |
3 |
1 |
34 |
388.531 |
8 |
↓
|
|
|
|
Analogs
-
602228
-
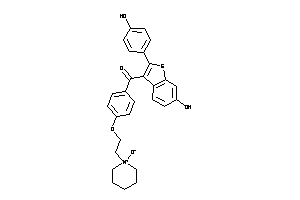
-
34537110
-
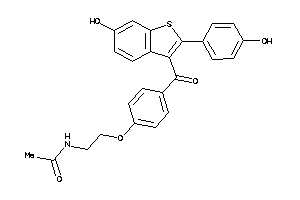
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2B-4-E |
Serotonin 2b (5-HT2b) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
69 |
0.29 |
Binding ≤ 10μM
|
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.37 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7 |
0.34 |
Binding ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.33 |
Binding ≤ 10μM
|
|
PLD1-1-E |
Phospholipase D1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8500 |
0.21 |
Binding ≤ 10μM
|
|
PLD2-1-E |
Phospholipase D2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.23 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
38 |
0.31 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.35 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.34 |
Functional ≤ 10μM
|
|
ADO-1-E |
Aldehyde Oxidase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.25 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
9900 |
0.21 |
ADME/T ≤ 10μM
|
|
ERG2-2-F |
C-8 Sterol Isomerase (cluster #2 Of 2), Fungal |
Fungi |
66 |
0.30 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3162 |
0.23 |
Functional ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
10000 |
0.21 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
2000 |
0.23 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
5 |
0.34 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
3 |
0.35 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
4 |
0.35 |
Functional ≤ 10μM
|
|
Z81068-1-O |
Ishikawa (Uterine Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
23 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.22 |
10.88 |
-44.71 |
3 |
5 |
1 |
71 |
474.602 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.22 |
8.67 |
-11.05 |
2 |
5 |
0 |
70 |
473.594 |
7 |
↓
|
|
|
|
Analogs
-
22058336
-
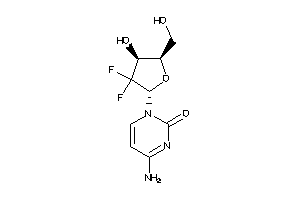
-
6532
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 62 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100515-1-O |
HPAC (Pancreatic Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
73 |
0.56 |
Functional ≤ 10μM |
|
Z100733-1-O |
CT26 (Colon Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
6 |
0.64 |
Functional ≤ 10μM |
|
Z100757-1-O |
SW1573 (Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
8300 |
0.40 |
Functional ≤ 10μM
|
|
Z103393-1-O |
SK-N-AS (cluster #1 Of 1), Other |
Other |
1100 |
0.46 |
Functional ≤ 10μM |
|
Z103407-1-O |
SW1990 (cluster #1 Of 2), Other |
Other |
1200 |
0.46 |
Functional ≤ 10μM
|
|
Z80048-1-O |
BXPC-3 (Pancreatic Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
160 |
0.53 |
Functional ≤ 10μM
|
|
Z80076-1-O |
CEM-SS (T-cell Leukemia) (cluster #1 Of 5), Other |
Other |
30 |
0.59 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
36 |
0.58 |
Functional ≤ 10μM
|
|
Z80132-1-O |
EL4 (Thymoma Cells) (cluster #1 Of 2), Other |
Other |
7 |
0.63 |
Functional ≤ 10μM |
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
330 |
0.50 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
43 |
0.57 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
746 |
0.48 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
7 |
0.63 |
Functional ≤ 10μM |
|
Z80224-9-O |
MCF7 (Breast Carcinoma Cells) (cluster #9 Of 14), Other |
Other |
80 |
0.55 |
Functional ≤ 10μM
|
|
Z80255-2-O |
MES-SA/DxS (Uterine Sarcoma Cells) (cluster #2 Of 2), Other |
Other |
9 |
0.63 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
40 |
0.58 |
Functional ≤ 10μM
|
|
Z80457-1-O |
SCC-25 (cluster #1 Of 1), Other |
Other |
47 |
0.57 |
Functional ≤ 10μM
|
|
Z80466-1-O |
SF268 (cluster #1 Of 4), Other |
Other |
10 |
0.62 |
Functional ≤ 10μM
|
|
Z80478-1-O |
SK-LU-1 (cluster #1 Of 1), Other |
Other |
55 |
0.56 |
Functional ≤ 10μM
|
|
Z80482-2-O |
SK-MEL-2 (Melanoma Cells) (cluster #2 Of 4), Other |
Other |
7110 |
0.40 |
Functional ≤ 10μM |
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
1700 |
0.45 |
Functional ≤ 10μM
|
|
Z80561-1-O |
U2OS (Osteosarcoma Cells) (cluster #1 Of 1), Other |
Other |
91 |
0.55 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
130 |
0.54 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
90 |
0.55 |
Functional ≤ 10μM
|
|
Z80702-1-O |
BG-1 Cell Line (cluster #1 Of 1), Other |
Other |
680 |
0.48 |
Functional ≤ 10μM
|
|
Z80726-1-O |
TRAMP-C1A (Prostate Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
9 |
0.63 |
Functional ≤ 10μM
|
|
Z80730-1-O |
C2D Cell Line (cluster #1 Of 1), Other |
Other |
12 |
0.62 |
Functional ≤ 10μM
|
|
Z80731-1-O |
C2G Cell Line (cluster #1 Of 1), Other |
Other |
4 |
0.65 |
Functional ≤ 10μM
|
|
Z80732-1-O |
TRAMP-C2H (Prostate Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
5 |
0.65 |
Functional ≤ 10μM
|
|
Z80742-1-O |
C6 (Glioma Cells) (cluster #1 Of 3), Other |
Other |
504 |
0.49 |
Functional ≤ 10μM |
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
130 |
0.54 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
10 |
0.62 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
8 |
0.63 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
35 |
0.58 |
Functional ≤ 10μM
|
|
Z81072-2-O |
Jurkat (Acute Leukemic T-cells) (cluster #2 Of 10), Other |
Other |
45 |
0.57 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
512 |
0.49 |
Functional ≤ 10μM |
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
8 |
0.63 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
245 |
0.51 |
Functional ≤ 10μM |
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
10 |
0.62 |
Functional ≤ 10μM
|
|
Z80335-1-O |
NHDF (cluster #1 Of 2), Other |
Other |
22 |
0.60 |
ADME/T ≤ 10μM
|
|
Z80704-1-O |
BJ (Foreskin Fibroblast Cells) (cluster #1 Of 2), Other |
Other |
9880 |
0.39 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.60 |
-4.98 |
-22.45 |
4 |
7 |
0 |
111 |
263.2 |
2 |
↓
|
|