We assess the chemical diversity of a subset by clustering the molecules. First, we sort ligands by increasing molecular weight. Then, we use the SUBSET 1.0 algorithm ( Voigt JH, Bienfait B, Wang S, Nicklaus MC. JCICS, 2001, 41, 702-12) to progressively select compounds that differ from those previously selected by at least the Tanimoto cutoff, using ChemAxon default fingerprints. The resulting representatives have two interesting properties:
| Tanimoto Cutoff Level | 60% | 70% | 80% | 90% | 100% |
|---|---|---|---|---|---|
| Number of Representatives | 267 | 464 | 922 | 2,358 | 11,581 |
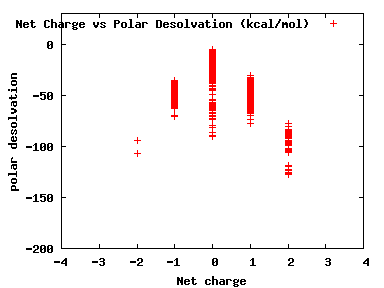
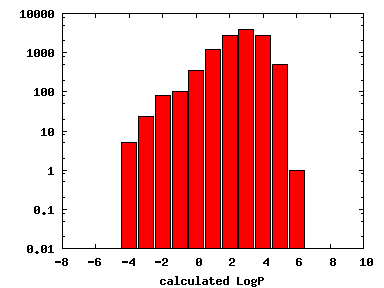
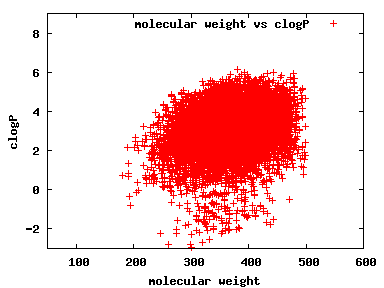
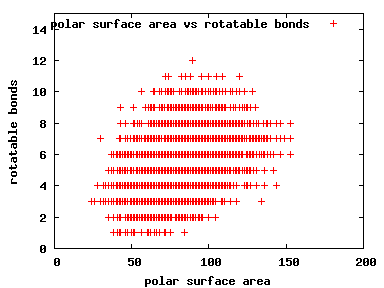
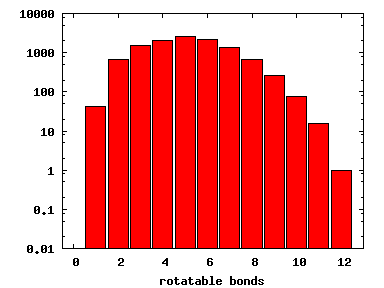
We compute the physical properties of each molecule in the subset, and graph them below.
Download Calculated Physical Properties

| Format | Reference(pH 7) | Mid(pH 6-8) | High(pH 8-9.5) | Low(pH 4.5-6) | Download Unix |
Download Windows |
|---|---|---|---|---|---|---|
| SMILES | All | All | All | All | ||
| MOL2 | All | All | All | All | Single Usual Metals All | Single Usual Metals All |
| SDF | All | All | All | All | Single Usual Metals All | Single Usual Metals All |
| Flexibase | Not Available | Not Available | Not Available | Not Available |