|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,4aR,5S,8aS)-5-[2-(5,8-diacetoxy-1,4-dihydronaphthalen-2-yl)ethyl]-1,4a-dimethyl-6-methylene-dec
[(1S,4aR,5S,8aS)-5-[2-(5,8-diace…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.05 |
19.05 |
-17.07 |
0 |
6 |
0 |
79 |
522.682 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,4aR,5S,8aR)-5-[2-(5,8-diacetoxy-1,4-dihydronaphthalen-2-yl)ethyl]-1,4a-dimethyl-6-methylene-dec
[(1S,4aR,5S,8aR)-5-[2-(5,8-diace…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.05 |
18.61 |
-17.42 |
0 |
6 |
0 |
79 |
522.682 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.07 |
13.63 |
-9.27 |
0 |
4 |
0 |
60 |
420.549 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.07 |
13.66 |
-9.88 |
0 |
4 |
0 |
60 |
420.549 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,4aR,5S,8aS)-5-[2-[(4aR,9aR)-9,10-dioxo-1,4,4a,9a-tetrahydroanthracen-2-yl]ethyl]-1,4a-dimethyl-
[(1S,4aR,5S,8aS)-5-[2-[(4aR,9aR)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.87 |
16.92 |
-10 |
0 |
4 |
0 |
60 |
488.668 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,4aR,5S,8aR)-5-[2-[(4aR,9aR)-9,10-dioxo-1,4,4a,9a-tetrahydroanthracen-2-yl]ethyl]-1,4a-dimethyl-
[(1S,4aR,5S,8aR)-5-[2-[(4aR,9aR)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.87 |
16.49 |
-10.29 |
0 |
4 |
0 |
60 |
488.668 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.27 |
15.58 |
-18.71 |
0 |
7 |
0 |
91 |
524.654 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.15 |
7.81 |
-16.81 |
2 |
5 |
0 |
79 |
440.58 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.15 |
7.78 |
-16.86 |
2 |
5 |
0 |
79 |
440.58 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(E,4R)-5-[(1S,3R,4aS,8aS)-3-hydroxy-5,5,8a-trimethyl-2-methylene-decalin-1-yl]-3-methyl-pent-2-ene-1
(E,4R)-5-[(1S,3R,4aS,8aS)-3-hydr…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
3.49 |
-4.24 |
3 |
3 |
0 |
61 |
322.489 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(E,4S)-5-[(1S,3R,4aS,8aS)-3-hydroxy-5,5,8a-trimethyl-2-methylene-decalin-1-yl]-3-methyl-pent-2-ene-1
(E,4S)-5-[(1S,3R,4aS,8aS)-3-hydr…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
2.39 |
-5.04 |
3 |
3 |
0 |
61 |
322.489 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(E,4R)-5-[(1R,3R,4aS,8aS)-3-hydroxy-5,5,8a-trimethyl-2-methylene-decalin-1-yl]-3-methyl-pent-2-ene-1
(E,4R)-5-[(1R,3R,4aS,8aS)-3-hydr…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
2.19 |
-5.87 |
3 |
3 |
0 |
61 |
322.489 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(E,4S)-5-[(1R,3R,4aS,8aS)-3-hydroxy-5,5,8a-trimethyl-2-methylene-decalin-1-yl]-3-methyl-pent-2-ene-1
(E,4S)-5-[(1R,3R,4aS,8aS)-3-hydr…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
2.1 |
-6.63 |
3 |
3 |
0 |
61 |
322.489 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(E,1R)-1-[[(1S,3R,4aS,8aS)-3-acetoxy-5,5,8a-trimethyl-2-methylene-decalin-1-yl]methyl]-4-acetoxy-2-
[(E,1R)-1-[[(1S,3R,4aS,8aS)-3-ac…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.98 |
14.41 |
-11.63 |
0 |
6 |
0 |
79 |
448.6 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(E,1S)-1-[[(1S,3R,4aS,8aS)-3-acetoxy-5,5,8a-trimethyl-2-methylene-decalin-1-yl]methyl]-4-acetoxy-2-
[(E,1S)-1-[[(1S,3R,4aS,8aS)-3-ac…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.98 |
14.71 |
-13.04 |
0 |
6 |
0 |
79 |
448.6 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(E,1R)-1-[[(1R,3R,4aS,8aS)-3-acetoxy-5,5,8a-trimethyl-2-methylene-decalin-1-yl]methyl]-4-acetoxy-2-
[(E,1R)-1-[[(1R,3R,4aS,8aS)-3-ac…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.98 |
14.37 |
-12.32 |
0 |
6 |
0 |
79 |
448.6 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(E,1S)-1-[[(1R,3R,4aS,8aS)-3-acetoxy-5,5,8a-trimethyl-2-methylene-decalin-1-yl]methyl]-4-acetoxy-2-
[(E,1S)-1-[[(1R,3R,4aS,8aS)-3-ac…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.98 |
14.81 |
-11.42 |
0 |
6 |
0 |
79 |
448.6 |
10 |
↓
|
|
|
|
Analogs
-
3994486
-
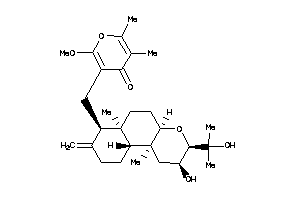
-
3994488
-
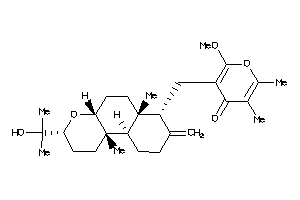
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.56 |
12.06 |
-10.39 |
1 |
4 |
0 |
60 |
442.64 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
1.48 |
-13.17 |
3 |
5 |
0 |
87 |
350.455 |
3 |
↓
|
|
|
|
Analogs
-
26145455
-
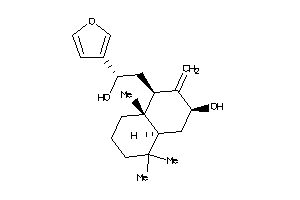
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.83 |
4.69 |
-5.66 |
2 |
3 |
0 |
54 |
318.457 |
3 |
↓
|
|
|
|
Analogs
-
26144778
-
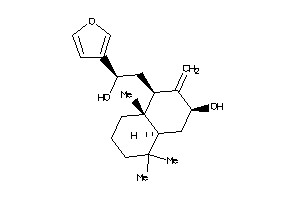
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.83 |
4.61 |
-5.4 |
2 |
3 |
0 |
54 |
318.457 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.34 |
11.55 |
-10.28 |
1 |
6 |
0 |
78 |
445.6 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.34 |
11.67 |
-11.59 |
1 |
6 |
0 |
78 |
445.6 |
11 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
3.29 |
-13.93 |
3 |
5 |
0 |
87 |
350.455 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
2-[(3S,4R)-4-[2-[(1R,4aS,5R,6R,8aS)-6-hydroxy-5-(hydroxymethyl)-5,8a-dimethyl-2-methylene-decalin-1-
2-[(3S,4R)-4-[2-[(1R,4aS,5R,6R,8…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
10.44 |
-57.27 |
2 |
8 |
-1 |
133 |
499.58 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
2-[(3S,4S)-4-[2-[(1R,4aS,5R,6R,8aS)-6-hydroxy-5-(hydroxymethyl)-5,8a-dimethyl-2-methylene-decalin-1-
2-[(3S,4S)-4-[2-[(1R,4aS,5R,6R,8…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
10.37 |
-56.93 |
2 |
8 |
-1 |
133 |
499.58 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(E)-5-[(1S,4aR,5R,6R,8aR)-5-hydroxy-5,6,8a-trimethyl-2-methylene-decalin-1-yl]-3-methyl-pent-2-enoic
(E)-5-[(1S,4aR,5R,6R,8aR)-5-hydr…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.46 |
8.89 |
-47.62 |
1 |
3 |
-1 |
60 |
319.465 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.46 |
6.86 |
-5.56 |
2 |
3 |
0 |
58 |
320.473 |
4 |
↓
|
|
|
|
Analogs
-
8763194
-
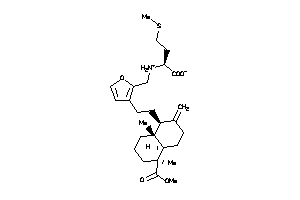
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.68 |
14.33 |
-38.08 |
2 |
6 |
0 |
96 |
491.694 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.75 |
14.21 |
-39.49 |
1 |
4 |
1 |
44 |
414.61 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.73 |
9.06 |
-51.02 |
3 |
4 |
1 |
67 |
360.518 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.35 |
13.07 |
-41.39 |
1 |
4 |
1 |
44 |
388.572 |
7 |
↓
|
|
|
|
Analogs
-
8917901
-
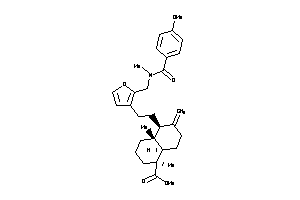
-
8917902
-
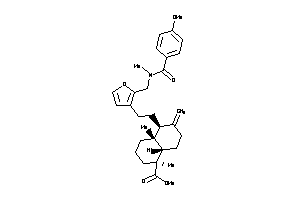
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.97 |
14.27 |
-12.4 |
0 |
6 |
0 |
69 |
507.671 |
9 |
↓
|
|
|
|
Analogs
-
32138309
-
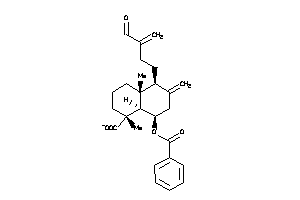
-
32138310
-
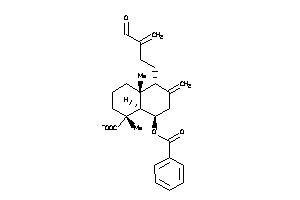
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.88 |
13.73 |
-58.37 |
0 |
5 |
-1 |
84 |
423.529 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.88 |
11.89 |
-12.27 |
1 |
5 |
0 |
81 |
424.537 |
8 |
↓
|
|
|
|
Analogs
-
32138309
-

-
32138310
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.88 |
14.26 |
-68.85 |
0 |
5 |
-1 |
84 |
423.529 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.88 |
12.33 |
-15.14 |
1 |
5 |
0 |
81 |
424.537 |
8 |
↓
|
|
|
|
Analogs
-
32138309
-

-
32138310
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.88 |
13.84 |
-47.4 |
0 |
5 |
-1 |
84 |
423.529 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.88 |
12.51 |
-13.37 |
1 |
5 |
0 |
81 |
424.537 |
8 |
↓
|
|
|
|
Analogs
-
9042927
-
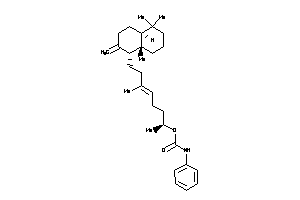
-
9042931
-
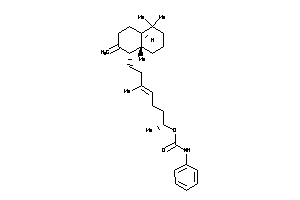
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(Z,1S)-7-[(1S,4aS,8aS)-5,5,8a-trimethyl-2-methylene-decalin-1-yl]-1,5-dimethyl-hept-4-enyl]
[(Z,1S)-7-[(1S,4aS,8aS)-5,5,8a-t…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.59 |
15.68 |
-5.67 |
1 |
3 |
0 |
38 |
451.695 |
9 |
↓
|
|
|
|
Analogs
-
9042927
-

-
9042931
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
[(Z,1R)-7-[(1S,4aS,8aS)-5,5,8a-trimethyl-2-methylene-decalin-1-yl]-1,5-dimethyl-hept-4-enyl]
[(Z,1R)-7-[(1S,4aS,8aS)-5,5,8a-t…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.59 |
15.7 |
-5.99 |
1 |
3 |
0 |
38 |
451.695 |
9 |
↓
|
|