|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.71 |
8.61 |
-55.1 |
2 |
5 |
-1 |
98 |
485.685 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.60 |
17.22 |
-113.5 |
0 |
7 |
-2 |
124 |
568.751 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.60 |
15.27 |
-60.62 |
1 |
7 |
-1 |
121 |
569.759 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.60 |
16.97 |
-112.98 |
0 |
7 |
-2 |
124 |
568.751 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.60 |
15 |
-59.03 |
1 |
7 |
-1 |
121 |
569.759 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.60 |
16.91 |
-111.81 |
0 |
7 |
-2 |
124 |
568.751 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.60 |
14.93 |
-50.98 |
1 |
7 |
-1 |
121 |
569.759 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,4aS,6aR,6aS,6bR,8aR,10S,12aS,14bR)-10-acetamido-2,4a,6a,6b,9,9,12a-heptamethyl-13-oxo-3,4,5,6,6a
(2S,4aS,6aR,6aS,6bR,8aR,10S,12aS…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHI1-2-E |
11-beta-hydroxysteroid Dehydrogenase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
825 |
0.23 |
Binding ≤ 10μM
|
|
DHI2-1-E |
11-beta-hydroxysteroid Dehydrogenase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
458 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.40 |
13.26 |
-71.09 |
1 |
5 |
-1 |
86 |
510.739 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.40 |
11.29 |
-20.64 |
2 |
5 |
0 |
83 |
511.747 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.68 |
15.14 |
-18.6 |
2 |
6 |
0 |
96 |
583.854 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.68 |
15.44 |
-54.48 |
3 |
6 |
1 |
97 |
584.862 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.92 |
13.7 |
-18.01 |
2 |
6 |
0 |
96 |
555.8 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.92 |
14.08 |
-56.64 |
3 |
6 |
1 |
97 |
556.808 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,4aS,6aR,6aS,6bR,8aR,10S,12aS,14bR)-10-(methanesulfonamido)-2,4a,6a,6b,9,9,12a-heptamethyl-13-oxo
(2S,4aS,6aR,6aS,6bR,8aR,10S,12aS…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.42 |
10.78 |
-73.28 |
1 |
6 |
-1 |
103 |
546.794 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.42 |
8.81 |
-24.6 |
2 |
6 |
0 |
101 |
547.802 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.84 |
15.56 |
-14.72 |
1 |
4 |
0 |
64 |
560.819 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,4aS,6aR,6aS,6bR,8aR,10S,12aS,14bR)-10-(2-aminoacetyl)oxy-2,4a,6a,6b,9,9,12a-heptamethyl-13-oxo-3
(2S,4aS,6aR,6aS,6bR,8aR,10S,12aS…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.92 |
12.65 |
-66.34 |
2 |
6 |
-1 |
110 |
526.738 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.92 |
13.03 |
-98.92 |
3 |
6 |
0 |
111 |
527.746 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.92 |
11.07 |
-56.49 |
4 |
6 |
1 |
108 |
528.754 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,4aS,6aR,6bS,8aR,10S,12aS,14aS,14bS)-10-hydroxy-2,4a,6a,6b,9,9,12a-heptamethyl-14-oxo-3,4,5,6,7,8
(2S,4aS,6aR,6bS,8aR,10S,12aS,14a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.62 |
11.61 |
-66.46 |
1 |
4 |
-1 |
77 |
469.686 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.62 |
9.61 |
-15.39 |
2 |
4 |
0 |
75 |
470.694 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3S,4aR,6aR,6bS,8aS,11S,12aR,14aR,14bS)-4,4,6a,6b,8a,11,14b-heptamethyl-14-oxo-11-ureido-2,3,4a,5,6
[(3S,4aR,6aR,6bS,8aS,11S,12aR,14…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHI1-2-E |
11-beta-hydroxysteroid Dehydrogenase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8300 |
0.19 |
Binding ≤ 10μM
|
|
DHI2-1-E |
11-beta-hydroxysteroid Dehydrogenase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
104 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.96 |
10.91 |
-15.46 |
3 |
6 |
0 |
98 |
526.762 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3S,4aR,6aR,6bS,8aS,11S,12aR,14aR,14bS)-11-(hydroxycarbamoylamino)-4,4,6a,6b,8a,11,14b-heptamethyl-
[(3S,4aR,6aR,6bS,8aS,11S,12aR,14…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.02 |
11.04 |
-19.71 |
3 |
7 |
0 |
105 |
542.761 |
3 |
↓
|
|
|
|
Analogs
-
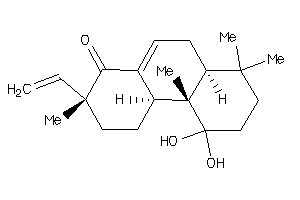
34820700
-
-
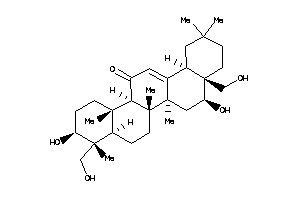
26292380
-
-
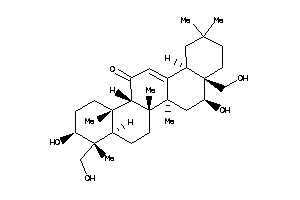
26292384
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aS,5S,6aR,6aS,6bR,8aS,9R,10S,12aS,14bR)-5,10-dihydroxy-4a,9-bis(hydroxymethyl)-2,2,6a,6b,9,12a-hex
(4aS,5S,6aR,6aS,6bR,8aS,9R,10S,1…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
3.4 |
-11.08 |
4 |
5 |
0 |
98 |
488.709 |
2 |
↓
|
|
|
|
Analogs
-
34820700
-

-
26292380
-

-
26292384
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aS,5S,6aS,6aS,6bR,8aS,9R,10S,12aS,14bR)-5,10-dihydroxy-4a,9-bis(hydroxymethyl)-2,2,6a,6b,9,12a-hex
(4aS,5S,6aS,6aS,6bR,8aS,9R,10S,1…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
4.92 |
-11.31 |
4 |
5 |
0 |
98 |
488.709 |
2 |
↓
|
|