|
|
Analogs
-
45329188
-
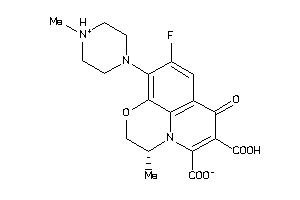
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.45 |
8.94 |
-80.57 |
2 |
9 |
0 |
116 |
405.382 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.45 |
6.6 |
-50.9 |
1 |
9 |
-1 |
115 |
404.374 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.20 |
8.68 |
-118.34 |
2 |
9 |
0 |
122 |
405.382 |
2 |
↓
|
|
|
|
Analogs
-
40620525
-
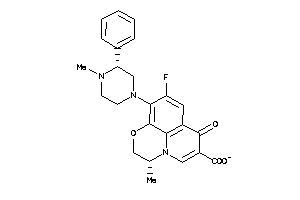
-
40620529
-
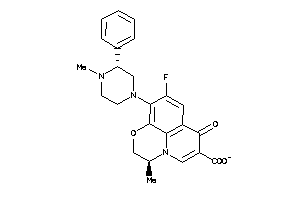
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.07 |
11.29 |
-100.08 |
2 |
7 |
0 |
91 |
423.444 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.07 |
9.27 |
-58.12 |
3 |
7 |
1 |
88 |
424.452 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.07 |
10.01 |
-63.72 |
1 |
7 |
-1 |
87 |
422.436 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.22 |
8.51 |
-60.85 |
2 |
15 |
1 |
179 |
611.611 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.22 |
6.14 |
-26.02 |
1 |
15 |
0 |
178 |
610.603 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.22 |
8.17 |
-60.25 |
2 |
15 |
1 |
179 |
611.611 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.22 |
5.8 |
-24.91 |
1 |
15 |
0 |
178 |
610.603 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.22 |
8.08 |
-55.27 |
2 |
15 |
1 |
179 |
611.611 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.22 |
5.71 |
-20.28 |
1 |
15 |
0 |
178 |
610.603 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.22 |
8.94 |
-60.43 |
2 |
15 |
1 |
179 |
611.611 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.22 |
6.57 |
-26.78 |
1 |
15 |
0 |
178 |
610.603 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.22 |
7.75 |
-54.89 |
2 |
15 |
1 |
179 |
611.611 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.22 |
5.38 |
-20.18 |
1 |
15 |
0 |
178 |
610.603 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.22 |
8.62 |
-65.93 |
2 |
15 |
1 |
179 |
611.611 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.22 |
6.26 |
-32.77 |
1 |
15 |
0 |
178 |
610.603 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.84 |
15.81 |
-49.57 |
2 |
7 |
1 |
68 |
529.721 |
13 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.84 |
13.47 |
-11.73 |
1 |
7 |
0 |
67 |
528.713 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.99 |
14.61 |
-58.34 |
1 |
9 |
1 |
92 |
490.552 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.99 |
12.27 |
-20.03 |
0 |
9 |
0 |
90 |
489.544 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.01 |
4.88 |
-65.17 |
3 |
11 |
-1 |
155 |
432.388 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.01 |
2.86 |
-17.62 |
4 |
11 |
0 |
152 |
433.396 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-3.75 |
2.99 |
-30.4 |
4 |
11 |
0 |
158 |
433.396 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.01 |
4.85 |
-65.49 |
3 |
11 |
-1 |
155 |
432.388 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.01 |
2.82 |
-17.85 |
4 |
11 |
0 |
152 |
433.396 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-3.75 |
2.97 |
-30.48 |
4 |
11 |
0 |
158 |
433.396 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.00 |
16.17 |
-58.3 |
1 |
9 |
1 |
92 |
518.606 |
12 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.00 |
13.83 |
-20 |
0 |
9 |
0 |
90 |
517.598 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
17.73 |
-58.19 |
1 |
9 |
1 |
92 |
546.66 |
14 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.01 |
15.39 |
-19.89 |
0 |
9 |
0 |
90 |
545.652 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.85 |
17.37 |
-49.54 |
2 |
7 |
1 |
68 |
557.775 |
15 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.85 |
15.04 |
-11.67 |
1 |
7 |
0 |
67 |
556.767 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.02 |
9.21 |
-69.09 |
2 |
11 |
-1 |
144 |
474.469 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.02 |
7.2 |
-22.37 |
3 |
11 |
0 |
141 |
475.477 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.73 |
7.37 |
-34.94 |
3 |
11 |
0 |
147 |
475.477 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.02 |
9.4 |
-66.79 |
2 |
11 |
-1 |
144 |
474.469 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.02 |
7.39 |
-19.69 |
3 |
11 |
0 |
141 |
475.477 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.73 |
7.51 |
-35.48 |
3 |
11 |
0 |
147 |
475.477 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.37 |
11.05 |
-53.09 |
1 |
7 |
1 |
65 |
390.435 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.37 |
8.72 |
-14.74 |
0 |
7 |
0 |
64 |
389.427 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.37 |
10.71 |
-52.35 |
1 |
7 |
1 |
65 |
390.435 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.37 |
8.38 |
-13.84 |
0 |
7 |
0 |
64 |
389.427 |
4 |
↓
|
|