|
|
Analogs
-
33816926
-
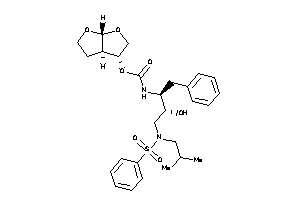
-
43989757
-
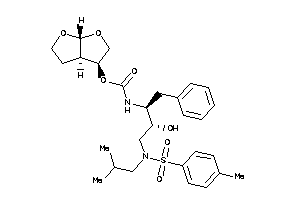
-
43989762
-
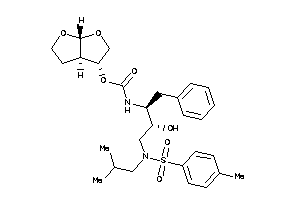
-
14946404
-
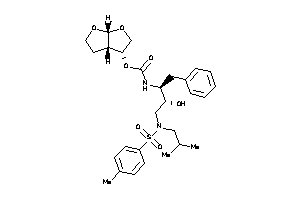
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3S,3aR,6aR)-2,3,3a,4,5,6a-hexahydrofuro[2,3-b]furan-3-yl]
[(3S,3aR,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.25 |
6.56 |
-15.72 |
2 |
9 |
0 |
114 |
532.659 |
12 |
↓
|
|
|
|
Analogs
-
43989757
-

-
43989762
-

-
33816925
-
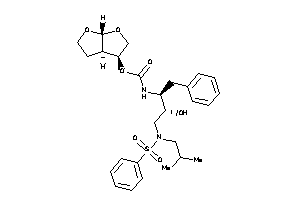
-
14946404
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3R,3aR,6aR)-2,3,3a,4,5,6a-hexahydrofuro[2,3-b]furan-3-yl]
[(3R,3aR,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.25 |
6.65 |
-15.85 |
2 |
9 |
0 |
114 |
532.659 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexahydrofuro[2,3-b]furan-3-yl]
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
162 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.75 |
13.65 |
-12.48 |
2 |
9 |
0 |
107 |
634.814 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexahydrofuro[2,3-b]furan-3-yl]
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
162 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.75 |
14.34 |
-11.78 |
2 |
9 |
0 |
107 |
634.814 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexahydrofuro[2,3-b]furan-3-yl]
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
268 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.96 |
14.35 |
-11.91 |
2 |
9 |
0 |
107 |
636.83 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexahydrofuro[2,3-b]furan-3-yl]
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
268 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.96 |
14.31 |
-11.36 |
2 |
9 |
0 |
107 |
636.83 |
9 |
↓
|
|
|
|
Analogs
-
26148870
-
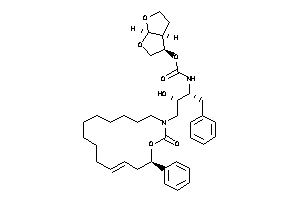
-
26148878
-
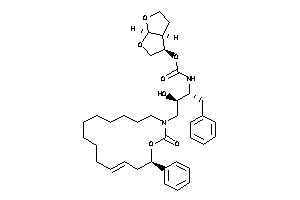
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexahydrofuro[2,3-b]furan-3-yl]
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
167 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.75 |
13.63 |
-12.52 |
2 |
9 |
0 |
107 |
634.814 |
9 |
↓
|
|
|
|
Analogs
-
26148870
-

-
26148878
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexahydrofuro[2,3-b]furan-3-yl]
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
167 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.75 |
14.19 |
-11.88 |
2 |
9 |
0 |
107 |
634.814 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexahydrofuro[2,3-b]furan-3-yl]
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
357 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.74 |
13.96 |
-12.25 |
2 |
9 |
0 |
107 |
622.803 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexahydrofuro[2,3-b]furan-3-yl]
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
189 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.46 |
12.58 |
-12.52 |
2 |
9 |
0 |
107 |
608.776 |
9 |
↓
|
|
|
|
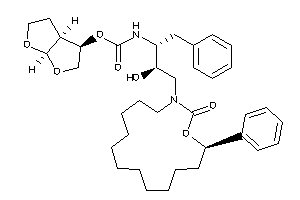
Analogs
-
26153404
-
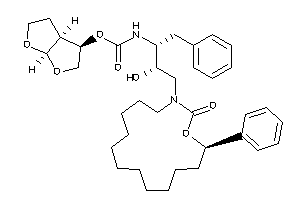
-
26153413
-
-
26158836
-
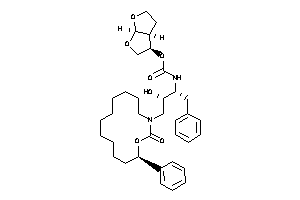
-
26158845
-
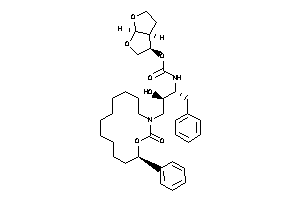
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexahydrofuro[5,4-b]furan-3-yl]
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
268 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.96 |
13.85 |
-11.17 |
2 |
9 |
0 |
107 |
636.83 |
9 |
↓
|
|
|
|
Analogs
-
26153404
-

-
26153413
-

-
26158836
-

-
26158845
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexahydrofuro[5,4-b]furan-3-yl]
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
268 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.96 |
14.23 |
-9.78 |
2 |
9 |
0 |
107 |
636.83 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexahydrofuro[5,4-b]furan-3-yl]
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.92 |
15.94 |
-15.33 |
2 |
9 |
0 |
107 |
648.841 |
22 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexahydrofuro[5,4-b]furan-3-yl]
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.92 |
16.75 |
-12.31 |
2 |
9 |
0 |
107 |
648.841 |
22 |
↓
|
|
|
|
Analogs
-
26146127
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexahydrofuro[5,4-b]furan-3-yl]
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.14 |
12.25 |
-14.62 |
2 |
9 |
0 |
107 |
572.743 |
21 |
↓
|
|
|
|
Analogs
-
26146127
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexahydrofuro[5,4-b]furan-3-yl]
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.14 |
13.06 |
-11.71 |
2 |
9 |
0 |
107 |
572.743 |
21 |
↓
|
|
|
|
Analogs
-
26146113
-

-
26146120
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexahydrofuro[5,4-b]furan-3-yl]
[(3R,3aS,6aR)-2,3,3a,4,5,6a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.14 |
13 |
-14.21 |
2 |
9 |
0 |
107 |
572.743 |
21 |
↓
|
|