|
|
Analogs
-
33819066
-
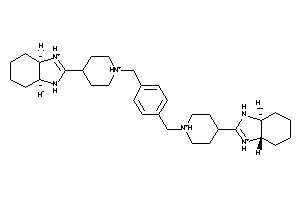
-
28112163
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-2-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #2 Of 3), Viral |
Viruses |
7000 |
0.19 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9_9HEPC |
A3EZI9
|
Hepatitis C Virus NS3 Protease/helicase, 9hepc |
7000 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.01 |
14.95 |
-238.11 |
6 |
6 |
4 |
61 |
520.81 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.57 |
15.14 |
-153.82 |
6 |
6 |
3 |
61 |
519.802 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.01 |
12.72 |
-141.67 |
5 |
6 |
3 |
60 |
519.802 |
6 |
↓
|
|
|
|
Analogs
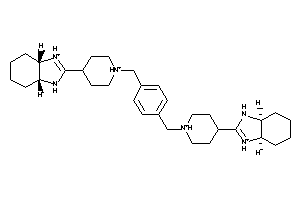
-
33819065
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-2-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #2 Of 3), Viral |
Viruses |
7000 |
0.19 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9_9HEPC |
A3EZI9
|
Hepatitis C Virus NS3 Protease/helicase, 9hepc |
7000 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.01 |
14.6 |
-238.05 |
6 |
6 |
4 |
61 |
520.81 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.57 |
14.71 |
-154.32 |
6 |
6 |
3 |
61 |
519.802 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.57 |
14.71 |
-153.35 |
6 |
6 |
3 |
61 |
519.802 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.29 |
6.85 |
-96.01 |
5 |
5 |
2 |
66 |
348.466 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.29 |
6.75 |
-100.32 |
5 |
5 |
2 |
66 |
348.466 |
4 |
↓
|
|
|
|
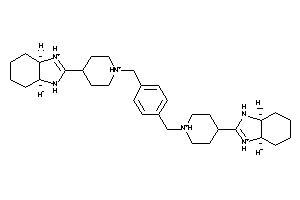
Analogs
-
1578004
-
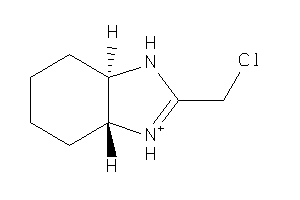
-
1578006
-
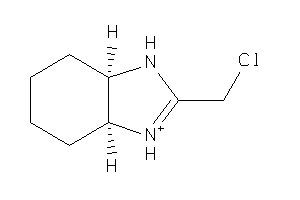
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
-3.01 |
-28.03 |
2 |
2 |
1 |
26 |
173.667 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
N-[[(3aS,7aR)-3a,4,5,6,7,7a-hexahydro-1H-benzimidazol-2-yl]carbamoyl-(4-chlorophenyl)carbamoyl]-4-ch
N-[[(3aS,7aR)-3a,4,5,6,7,7a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
11.37 |
-64.36 |
2 |
8 |
-1 |
109 |
473.34 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.67 |
12.79 |
-21.94 |
3 |
8 |
0 |
103 |
474.348 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.67 |
11.67 |
-51.21 |
2 |
8 |
-1 |
105 |
473.34 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
N-[[(3aS,7aS)-3a,4,5,6,7,7a-hexahydro-1H-benzimidazol-2-yl]carbamoyl-(4-chlorophenyl)carbamoyl]-4-ch
N-[[(3aS,7aS)-3a,4,5,6,7,7a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
11.21 |
-63.58 |
2 |
8 |
-1 |
109 |
473.34 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.67 |
12.33 |
-20.48 |
3 |
8 |
0 |
103 |
474.348 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
N-[[(3aR,7aR)-3a,4,5,6,7,7a-hexahydro-1H-benzimidazol-2-yl]carbamoyl-(4-chlorophenyl)carbamoyl]-4-ch
N-[[(3aR,7aR)-3a,4,5,6,7,7a-hexa…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
11.15 |
-63.41 |
2 |
8 |
-1 |
109 |
473.34 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.67 |
12.4 |
-20.73 |
3 |
8 |
0 |
103 |
474.348 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
14244894
-
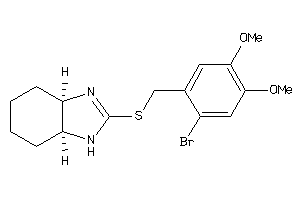
-
14244895
-
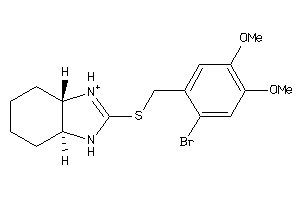
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.76 |
6.33 |
-6.59 |
1 |
4 |
0 |
43 |
385.327 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
14248098
-
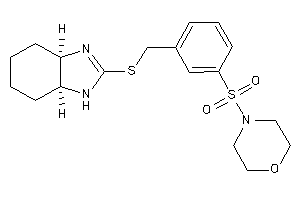
-
14248101
-
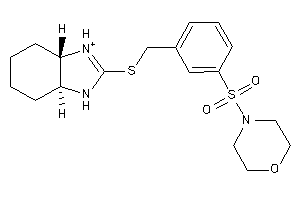
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
4.18 |
-10.91 |
1 |
6 |
0 |
71 |
395.55 |
5 |
↓
|
|