|
|
Analogs
-
13524658
-
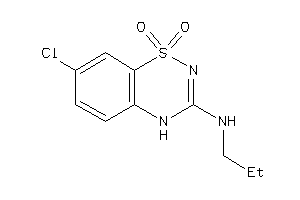
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
7.27 |
-21.35 |
2 |
5 |
0 |
71 |
335.816 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.05 |
6.82 |
-37.69 |
1 |
5 |
-1 |
73 |
334.808 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
5.55 |
-61.82 |
3 |
8 |
1 |
96 |
459.592 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
5.98 |
-19.13 |
2 |
5 |
0 |
71 |
281.381 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
4.8 |
-17.77 |
2 |
5 |
0 |
71 |
308.19 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.73 |
4.36 |
-35.03 |
1 |
5 |
-1 |
73 |
307.182 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.22 |
4.04 |
-17.8 |
2 |
5 |
0 |
71 |
294.163 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.22 |
3.59 |
-35.15 |
1 |
5 |
-1 |
73 |
293.155 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.21 |
4.95 |
-17.66 |
2 |
5 |
0 |
71 |
306.174 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.21 |
4.5 |
-34.95 |
1 |
5 |
-1 |
73 |
305.166 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.98 |
2.87 |
-20.34 |
2 |
6 |
0 |
80 |
287.316 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.17 |
4.58 |
-19.67 |
2 |
5 |
0 |
71 |
271.317 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.84 |
4.29 |
-20.92 |
2 |
6 |
0 |
83 |
306.322 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.84 |
4.75 |
-57.37 |
3 |
6 |
1 |
85 |
307.33 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.89 |
4.26 |
-20.8 |
2 |
6 |
0 |
83 |
306.322 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.89 |
4.73 |
-57.2 |
3 |
6 |
1 |
85 |
307.33 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.96 |
4.21 |
-20.43 |
2 |
6 |
0 |
83 |
306.322 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.96 |
4.62 |
-54.95 |
3 |
6 |
1 |
85 |
307.33 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
N-[3-[(3S,4aS,7aR)-1-methyl-2,4-dioxo-5,6,7,7a-tetrahydro-4aH-cyclopenta[b]pyridin-3-yl]-1,1-dioxo-4
N-[3-[(3S,4aS,7aR)-1-methyl-2,4-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.62 |
0.27 |
-107.21 |
1 |
10 |
-2 |
150 |
438.487 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.04 |
-0.19 |
-53.7 |
1 |
10 |
-1 |
144 |
439.495 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.04 |
0.42 |
-29.46 |
1 |
10 |
0 |
144 |
439.495 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
N-[3-[(3S,4aR,7aR)-1-methyl-2,4-dioxo-5,6,7,7a-tetrahydro-4aH-cyclopenta[b]pyridin-3-yl]-1,1-dioxo-4
N-[3-[(3S,4aR,7aR)-1-methyl-2,4-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.62 |
0.12 |
-105.49 |
1 |
10 |
-2 |
150 |
438.487 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.04 |
0.53 |
-28.28 |
1 |
10 |
0 |
144 |
439.495 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.04 |
0.23 |
-22.97 |
2 |
10 |
0 |
142 |
440.503 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
3.8 |
-28.61 |
0 |
7 |
0 |
100 |
293.304 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.64 |
1.4 |
-48.95 |
1 |
7 |
-1 |
93 |
337.425 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.64 |
1.79 |
-31.98 |
2 |
7 |
0 |
91 |
338.433 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.64 |
3.95 |
-68.88 |
3 |
7 |
1 |
92 |
339.441 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
4.57 |
-26.2 |
2 |
5 |
0 |
79 |
348.449 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.15 |
4.16 |
-43.73 |
1 |
5 |
-1 |
81 |
347.441 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
4.6 |
-26.01 |
2 |
5 |
0 |
79 |
348.449 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.15 |
4.2 |
-43.62 |
1 |
5 |
-1 |
81 |
347.441 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.95 |
2.99 |
-27.24 |
1 |
6 |
0 |
85 |
345.405 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.95 |
2.58 |
-43.58 |
0 |
6 |
-1 |
87 |
344.397 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50643-2-O |
Hepatitis C Virus (cluster #2 Of 5), Other |
Other |
38 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.07 |
4.26 |
-111.37 |
1 |
9 |
-2 |
147 |
515.613 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.63 |
5.46 |
-52.44 |
2 |
9 |
-1 |
145 |
516.621 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.04 |
4.93 |
-23.23 |
2 |
9 |
0 |
139 |
517.629 |
6 |
↓
|
|
|
|
Analogs
-
53122830
-
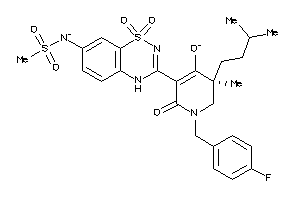
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
6.15 |
-111.32 |
1 |
10 |
-2 |
150 |
562.645 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.21 |
5.8 |
-219.43 |
0 |
10 |
-3 |
152 |
561.637 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.63 |
6.27 |
-61.01 |
1 |
10 |
-1 |
144 |
563.653 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
5.17 |
-53.13 |
1 |
10 |
-1 |
144 |
539.7 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.55 |
5.31 |
-220.08 |
0 |
10 |
-3 |
152 |
537.684 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.97 |
4.68 |
-50.74 |
1 |
10 |
-1 |
144 |
539.7 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
5.19 |
-54.88 |
1 |
10 |
-1 |
144 |
539.7 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.55 |
5.19 |
-217.05 |
0 |
10 |
-3 |
152 |
537.684 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.97 |
4.71 |
-50.02 |
1 |
10 |
-1 |
144 |
539.7 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
3.86 |
-54.27 |
1 |
10 |
-1 |
144 |
509.63 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.29 |
3.21 |
-222.63 |
0 |
10 |
-3 |
152 |
507.614 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.29 |
3.52 |
-111.64 |
1 |
10 |
-2 |
150 |
508.622 |
6 |
↓
|
|
|
|
Analogs
-
53122844
-
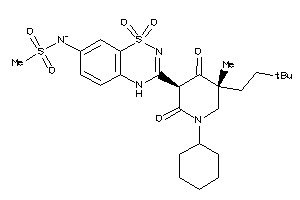
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
3.87 |
-53.29 |
1 |
10 |
-1 |
144 |
509.63 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.29 |
3.22 |
-221.94 |
0 |
10 |
-3 |
152 |
507.614 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.29 |
3.6 |
-111.11 |
1 |
10 |
-2 |
150 |
508.622 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
3.86 |
-53.98 |
1 |
10 |
-1 |
144 |
509.63 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.29 |
3.22 |
-221.84 |
0 |
10 |
-3 |
152 |
507.614 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.70 |
3.45 |
-116.43 |
0 |
10 |
-2 |
146 |
508.622 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
3.65 |
-53.89 |
1 |
10 |
-1 |
144 |
509.63 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.29 |
3.2 |
-222.42 |
0 |
10 |
-3 |
152 |
507.614 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.29 |
3.51 |
-111.44 |
1 |
10 |
-2 |
150 |
508.622 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
6.21 |
-109.51 |
1 |
10 |
-2 |
150 |
562.645 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.10 |
5.93 |
-217.2 |
0 |
10 |
-3 |
152 |
561.637 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.52 |
5.72 |
-51.8 |
1 |
10 |
-1 |
144 |
563.653 |
8 |
↓
|
|
|
|
Analogs
-
53122830
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.46 |
6.68 |
-112.42 |
1 |
10 |
-2 |
150 |
576.672 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.46 |
6.32 |
-220.48 |
0 |
10 |
-3 |
152 |
575.664 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.88 |
6.18 |
-54.72 |
1 |
10 |
-1 |
144 |
577.68 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
3.57 |
-109.55 |
1 |
11 |
-2 |
153 |
537.664 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.31 |
3.27 |
-219.01 |
0 |
11 |
-3 |
155 |
536.656 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.72 |
3.29 |
-54.4 |
1 |
11 |
-1 |
147 |
538.672 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
3.52 |
-105.91 |
1 |
11 |
-2 |
153 |
537.664 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.31 |
3.16 |
-213.76 |
0 |
11 |
-3 |
155 |
536.656 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.72 |
3.64 |
-58.87 |
1 |
11 |
-1 |
147 |
538.672 |
7 |
↓
|
|
|
|
Analogs
-
53119672
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
6.09 |
-113.46 |
1 |
10 |
-2 |
150 |
560.629 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.24 |
5.72 |
-221.32 |
0 |
10 |
-3 |
152 |
559.621 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.24 |
6.05 |
-57.94 |
2 |
10 |
-1 |
148 |
561.637 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.59 |
2.83 |
-52.27 |
1 |
10 |
-1 |
144 |
483.592 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.17 |
2.66 |
-213.88 |
0 |
10 |
-3 |
152 |
481.576 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.59 |
2.4 |
-48.78 |
1 |
10 |
-1 |
144 |
483.592 |
6 |
↓
|
|
|
|
Analogs
-
53122849
-
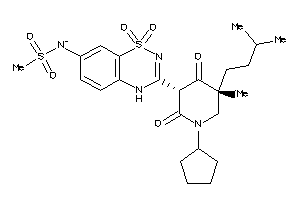
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
4.95 |
-51.21 |
1 |
10 |
-1 |
144 |
523.657 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.05 |
4.44 |
-216.41 |
0 |
10 |
-3 |
152 |
521.641 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.05 |
4.74 |
-108.24 |
1 |
10 |
-2 |
150 |
522.649 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
200 |
0.33 |
Binding ≤ 10μM
|
|
Q8JXU8-1-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #1 Of 1), Viral |
Viruses |
444 |
0.32 |
Functional ≤ 10μM
|
|
Z80169-1-O |
Huh-7 (Hepatocellular Carcinoma) (cluster #1 Of 1), Other |
Other |
444 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
6.59 |
-132.83 |
0 |
7 |
-2 |
106 |
395.44 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
21 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.54 |
7.48 |
-133.81 |
0 |
7 |
-2 |
106 |
423.494 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
80 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.52 |
7.37 |
-132.99 |
0 |
7 |
-2 |
106 |
409.467 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
65 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
7.74 |
-133.55 |
0 |
7 |
-2 |
106 |
423.494 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
65 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
7.75 |
-133.48 |
0 |
7 |
-2 |
106 |
423.494 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.22 |
5.61 |
-133.7 |
0 |
7 |
-2 |
106 |
379.397 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
150 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.47 |
6.47 |
-134.4 |
0 |
7 |
-2 |
106 |
393.424 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
289 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.25 |
6.87 |
-135.95 |
0 |
7 |
-2 |
106 |
407.451 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
90 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.49 |
6.37 |
-133.09 |
0 |
7 |
-2 |
106 |
393.424 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
118 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.00 |
6.33 |
-131.37 |
0 |
7 |
-2 |
106 |
391.408 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.00 |
6.65 |
-56.01 |
1 |
7 |
-1 |
104 |
392.416 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
179 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
7.11 |
-133.6 |
0 |
7 |
-2 |
106 |
405.435 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
78 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.44 |
6.55 |
-134.04 |
0 |
7 |
-2 |
106 |
393.424 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
41 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
7.15 |
-134.55 |
0 |
7 |
-2 |
106 |
407.451 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
41 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
7.23 |
-134.68 |
0 |
7 |
-2 |
106 |
407.451 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
41 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
7.28 |
-132.33 |
0 |
7 |
-2 |
106 |
407.451 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
41 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
7.22 |
-132.42 |
0 |
7 |
-2 |
106 |
407.451 |
3 |
↓
|
|