|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.49 |
9.51 |
-111.96 |
0 |
9 |
-2 |
129 |
527.001 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.49 |
8.64 |
-65.45 |
1 |
9 |
-1 |
127 |
528.009 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.49 |
9.56 |
-109.07 |
0 |
9 |
-2 |
129 |
527.001 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.49 |
8.68 |
-63.08 |
1 |
9 |
-1 |
127 |
528.009 |
8 |
↓
|
|
|
|
Analogs
-
16915212
-
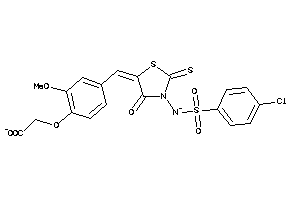
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
9.84 |
-110.33 |
0 |
9 |
-2 |
129 |
527.001 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.51 |
8.96 |
-64.1 |
1 |
9 |
-1 |
127 |
528.009 |
9 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
10.42 |
-112.23 |
0 |
9 |
-2 |
129 |
541.028 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.87 |
9.54 |
-65.75 |
1 |
9 |
-1 |
127 |
542.036 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
10.43 |
-109.37 |
0 |
9 |
-2 |
129 |
541.028 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.87 |
9.55 |
-63.56 |
1 |
9 |
-1 |
127 |
542.036 |
9 |
↓
|
|
|
|
Analogs
-
16912781
-
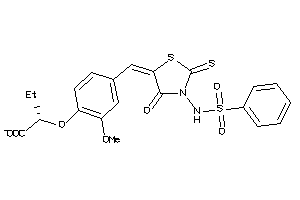
-
16912777
-
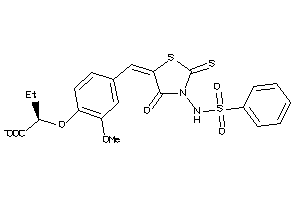
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.69 |
9.9 |
-73.28 |
1 |
9 |
-1 |
127 |
521.618 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.69 |
10.79 |
-129.09 |
0 |
9 |
-2 |
129 |
520.61 |
10 |
↓
|
|
|
|
Analogs
-
16912781
-

-
16912777
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.69 |
9.86 |
-66.05 |
1 |
9 |
-1 |
127 |
521.618 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.69 |
10.74 |
-121.22 |
0 |
9 |
-2 |
129 |
520.61 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.14 |
10.58 |
-73.04 |
1 |
9 |
-1 |
127 |
535.645 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.14 |
11.46 |
-129.51 |
0 |
9 |
-2 |
129 |
534.637 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.14 |
10.53 |
-65.83 |
1 |
9 |
-1 |
127 |
535.645 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.14 |
11.42 |
-121.47 |
0 |
9 |
-2 |
129 |
534.637 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
11.31 |
-122.63 |
0 |
9 |
-2 |
129 |
555.055 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.37 |
10.43 |
-70.19 |
1 |
9 |
-1 |
127 |
556.063 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
11.26 |
-114.7 |
0 |
9 |
-2 |
129 |
555.055 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.37 |
10.39 |
-63.6 |
1 |
9 |
-1 |
127 |
556.063 |
10 |
↓
|
|
|
|
Analogs
-
16912073
-
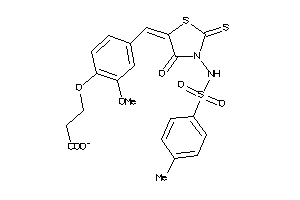
-
16919787
-
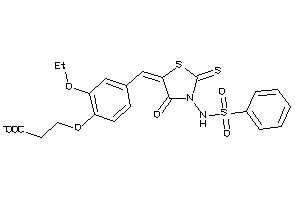
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.55 |
9.39 |
-62.39 |
1 |
9 |
-1 |
127 |
521.618 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.55 |
10.28 |
-115.67 |
0 |
9 |
-2 |
129 |
520.61 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
10.12 |
-109.31 |
0 |
9 |
-2 |
129 |
541.028 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.78 |
9.24 |
-60.23 |
1 |
9 |
-1 |
127 |
542.036 |
10 |
↓
|
|
|
|
Analogs
-
1300993
-
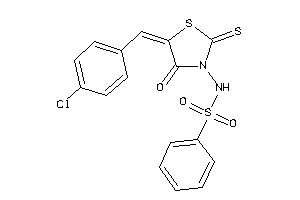
-
6142027
-
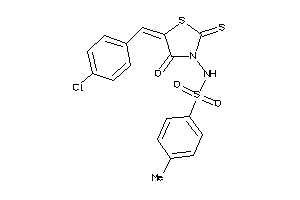
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
1500 |
0.33 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8_9HEPC |
Q8JXU8
|
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase, 9hepc |
1500 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
8.19 |
-60.17 |
0 |
5 |
-1 |
70 |
409.921 |
4 |
↓
|
|
|
|
Analogs
-
5378115
-
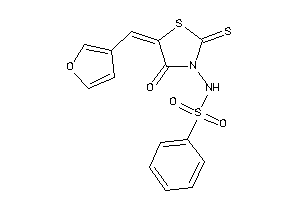
-
1214149
-
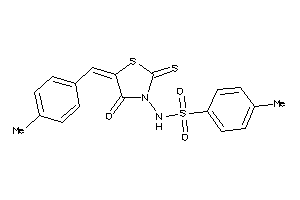
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
10000 |
0.28 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8_9HEPC |
Q8JXU8
|
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase, 9hepc |
10000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.01 |
7.22 |
-13.97 |
1 |
5 |
0 |
68 |
390.511 |
4 |
↓
|
|
|
|
Analogs
-
3964806
-
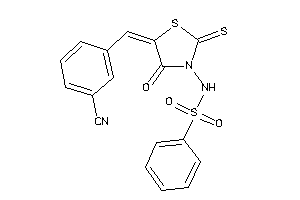
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
7.21 |
-13.7 |
1 |
5 |
0 |
68 |
390.511 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.98 |
8.09 |
-61.22 |
0 |
5 |
-1 |
70 |
389.503 |
4 |
↓
|
|
|
|
Analogs
-
15986554
-
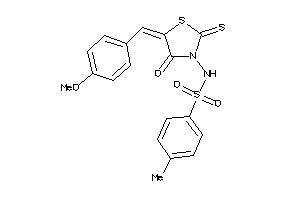
-
1300997
-
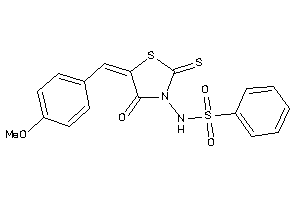
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
5000 |
0.29 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8_9HEPC |
Q8JXU8
|
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase, 9hepc |
5000 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
5.85 |
-15.29 |
1 |
6 |
0 |
77 |
406.51 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.62 |
6.74 |
-62.41 |
0 |
6 |
-1 |
79 |
405.502 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
6500 |
0.29 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8_9HEPC |
Q8JXU8
|
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase, 9hepc |
6500 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.68 |
6.84 |
-14.61 |
1 |
5 |
0 |
68 |
394.474 |
4 |
↓
|
|
|
|
Analogs
-
5378115
-

-
1231002
-
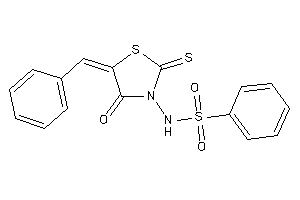
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
900 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
7.18 |
-20.92 |
1 |
6 |
0 |
92 |
401.494 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
1000 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.46 |
7.76 |
-17.61 |
1 |
5 |
0 |
68 |
444.481 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.46 |
8.68 |
-60.03 |
0 |
5 |
-1 |
70 |
443.473 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
1000 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
8.24 |
-57.39 |
0 |
5 |
-1 |
70 |
427.911 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.33 |
7.33 |
-16.58 |
1 |
5 |
0 |
68 |
428.919 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
200 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.85 |
8.65 |
-57.25 |
0 |
5 |
-1 |
70 |
444.366 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.85 |
7.73 |
-16 |
1 |
5 |
0 |
68 |
445.374 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
3000 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8_9HEPC |
Q8JXU8
|
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase, 9hepc |
3000 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
6.91 |
-13.9 |
1 |
5 |
0 |
68 |
412.464 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.82 |
7.81 |
-55.9 |
0 |
5 |
-1 |
70 |
411.456 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
1700 |
0.31 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8_9HEPC |
Q8JXU8
|
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase, 9hepc |
1700 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
7.81 |
-58.83 |
0 |
5 |
-1 |
70 |
411.456 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.82 |
6.9 |
-14.6 |
1 |
5 |
0 |
68 |
412.464 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
1900 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8_9HEPC |
Q8JXU8
|
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase, 9hepc |
1900 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
7.87 |
-54.48 |
0 |
5 |
-1 |
70 |
429.446 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.91 |
6.96 |
-15.28 |
1 |
5 |
0 |
68 |
430.454 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
1000 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
7.86 |
-59.07 |
0 |
5 |
-1 |
70 |
429.446 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.91 |
6.95 |
-19.03 |
1 |
5 |
0 |
68 |
430.454 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
200 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
7.87 |
-56.03 |
0 |
5 |
-1 |
70 |
429.446 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.91 |
6.96 |
-14.55 |
1 |
5 |
0 |
68 |
430.454 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8-2-V |
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase (cluster #2 Of 2), Viral |
Viruses |
8000 |
0.27 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8JXU8_9HEPC |
Q8JXU8
|
Hepatitis C Virus NS5B RNA-dependent RNA Polymerase, 9hepc |
8000 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.39 |
7.76 |
-13.69 |
1 |
5 |
0 |
68 |
404.538 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.39 |
8.65 |
-61.5 |
0 |
5 |
-1 |
70 |
403.53 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
7.8 |
-16.1 |
1 |
5 |
0 |
68 |
463.364 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
7.69 |
-20.54 |
1 |
5 |
0 |
68 |
463.364 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.29 |
8.41 |
-18.19 |
1 |
5 |
0 |
68 |
459.401 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
7.03 |
-19.6 |
1 |
6 |
0 |
77 |
475.4 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.90 |
7.03 |
-18.15 |
1 |
6 |
0 |
77 |
475.4 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.52 |
8.25 |
-15.99 |
1 |
5 |
0 |
68 |
479.819 |
4 |
↓
|
|