|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
43609325
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
8.36 |
-5.59 |
0 |
2 |
0 |
30 |
218.296 |
2 |
↓
|
|
|
|
Analogs
-
43609322
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
8.33 |
-5.91 |
0 |
2 |
0 |
30 |
218.296 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
1.55 |
-57.45 |
2 |
6 |
-1 |
102 |
365.475 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.85 |
3.49 |
-14.03 |
3 |
6 |
0 |
95 |
366.483 |
3 |
↓
|
|
|
|
Analogs
-
3382325
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
-1.18 |
-13.95 |
2 |
5 |
0 |
69 |
309.366 |
5 |
↓
|
|
|
|
Analogs
-
3382325
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
-0.67 |
-13.63 |
2 |
5 |
0 |
69 |
309.366 |
5 |
↓
|
|
|
|
Analogs
-
36747140
-
-
43413231
-
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
8.53 |
-67.37 |
1 |
4 |
-1 |
69 |
278.372 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.97 |
7.39 |
-13.29 |
2 |
4 |
0 |
66 |
279.38 |
4 |
↓
|
|
|
|
Analogs
-
36747140
-

-
43413231
-

Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
7.04 |
-44.28 |
1 |
4 |
-1 |
69 |
278.372 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.97 |
5.91 |
-9.54 |
2 |
4 |
0 |
66 |
279.38 |
4 |
↓
|
|
|
|
Analogs
-
575978
-
-
575980
-
-
575984
-
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.00 |
-0.92 |
-59.73 |
1 |
4 |
-1 |
69 |
300.378 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.00 |
-1.11 |
-45.86 |
1 |
4 |
-1 |
69 |
300.378 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.43 |
-0.36 |
-62.29 |
1 |
5 |
-1 |
78 |
288.323 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.43 |
-0.58 |
-45.83 |
1 |
5 |
-1 |
78 |
288.323 |
4 |
↓
|
|
|
|
Analogs
-
37833202
-
-
37833203
-
-
37833204
-
-
37833205
-
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.14 |
-0.72 |
-47.66 |
1 |
5 |
-1 |
78 |
302.35 |
5 |
↓
|
|
|
|
Analogs
-
37833202
-

-
37833203
-

-
37833204
-

-
37833205
-

Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.14 |
-1.36 |
-46.85 |
1 |
5 |
-1 |
78 |
302.35 |
5 |
↓
|
|
|
|
Analogs
-
53148846
-
-
53148848
-
-
53148851
-
-
53148854
-
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
-0.89 |
-44 |
1 |
4 |
-1 |
69 |
306.769 |
4 |
↓
|
|
|
|
Analogs
-
53148846
-

-
53148848
-

-
53148851
-

-
53148854
-

Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
-1.48 |
-44.9 |
1 |
4 |
-1 |
69 |
306.769 |
4 |
↓
|
|
|
|
Analogs
-
16551822
-
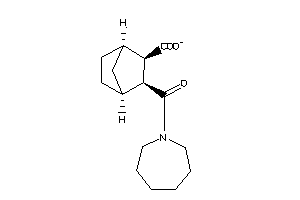
-
16551825
-
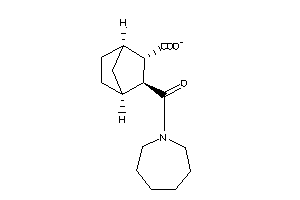
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.34 |
0.37 |
-62.33 |
0 |
4 |
-1 |
60 |
264.345 |
2 |
↓
|
|
|
|
Analogs
-
16551822
-

-
16551825
-

Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.34 |
0.09 |
-47.25 |
0 |
4 |
-1 |
60 |
264.345 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
0.8 |
-47.29 |
1 |
4 |
-1 |
69 |
290.314 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
0.42 |
-45.66 |
1 |
4 |
-1 |
69 |
290.314 |
3 |
↓
|
|
|
|
Analogs
-
8009553
-
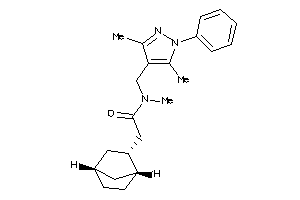
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
1.87 |
-10.71 |
0 |
4 |
0 |
38 |
351.494 |
5 |
↓
|
|
|
|
Analogs
-
8009551
-
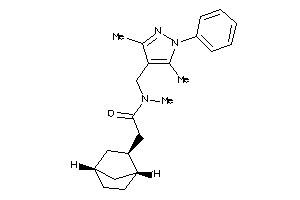
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
1.98 |
-10.14 |
0 |
4 |
0 |
38 |
351.494 |
5 |
↓
|
|
|
|
Analogs
-
8010005
-
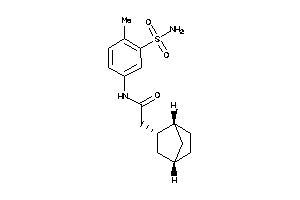
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.37 |
3.51 |
-18.19 |
3 |
5 |
0 |
89 |
322.43 |
4 |
↓
|
|
|
|
Analogs
-
8010003
-
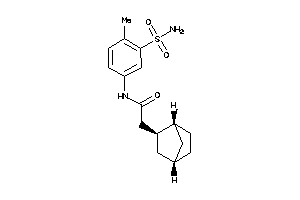
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.37 |
3.44 |
-13.07 |
3 |
5 |
0 |
89 |
322.43 |
4 |
↓
|
|
|
|
Analogs
-
8271516
-
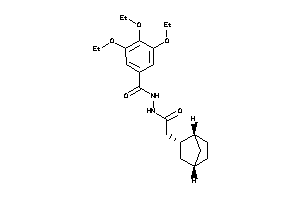
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
-1.7 |
-11.01 |
2 |
7 |
0 |
85 |
404.507 |
10 |
↓
|
|
|
|
Analogs
-
8271513
-
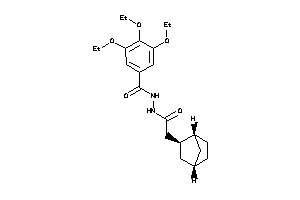
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
-1.73 |
-10.66 |
2 |
7 |
0 |
85 |
404.507 |
10 |
↓
|
|
|
|
Analogs
-
8486544
-
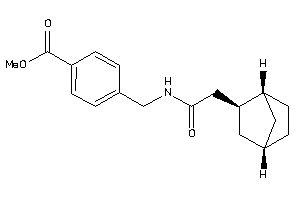
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.17 |
8.47 |
-9.92 |
1 |
4 |
0 |
55 |
301.386 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.27 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.21 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
10.13 |
-16.76 |
1 |
7 |
0 |
95 |
512.672 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
48 |
0.28 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2900 |
0.21 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2900 |
0.21 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7500 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
13.47 |
-20.93 |
1 |
7 |
0 |
84 |
524.731 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.53 |
13.95 |
-38.06 |
2 |
7 |
1 |
86 |
525.739 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.26 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.22 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
8.06 |
-43.26 |
5 |
7 |
1 |
125 |
516.728 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.06 |
7.75 |
-18.6 |
4 |
7 |
0 |
124 |
515.72 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.26 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.22 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
8.47 |
-42.78 |
5 |
7 |
1 |
125 |
516.728 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.06 |
8.23 |
-18.56 |
4 |
7 |
0 |
124 |
515.72 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.27 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.21 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.21 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.98 |
3.12 |
-57.63 |
5 |
9 |
1 |
148 |
582.809 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.98 |
2.79 |
-29.54 |
4 |
9 |
0 |
147 |
581.801 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.27 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.21 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.21 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.98 |
3.08 |
-57.51 |
5 |
9 |
1 |
148 |
582.809 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.98 |
2.76 |
-27.75 |
4 |
9 |
0 |
147 |
581.801 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.27 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.21 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.21 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.76 |
13.09 |
-22.9 |
2 |
11 |
0 |
136 |
608.765 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.58 |
12.01 |
-50.87 |
1 |
11 |
-1 |
142 |
607.757 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.25 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.19 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.19 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
8.35 |
-56.27 |
3 |
8 |
1 |
117 |
578.821 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.57 |
7.09 |
-24.43 |
2 |
8 |
0 |
113 |
577.813 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.25 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.19 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.19 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
8.31 |
-57.6 |
3 |
8 |
1 |
117 |
578.821 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.57 |
6.95 |
-23.57 |
2 |
8 |
0 |
113 |
577.813 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.25 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.19 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.19 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
8.3 |
-60.33 |
3 |
8 |
1 |
117 |
578.821 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.57 |
7.09 |
-29.72 |
2 |
8 |
0 |
113 |
577.813 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.25 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.19 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.19 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
8.16 |
-54.18 |
3 |
8 |
1 |
117 |
578.821 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.57 |
6.78 |
-24.25 |
2 |
8 |
0 |
113 |
577.813 |
6 |
↓
|
|
|
|
Analogs
-
3915327
-
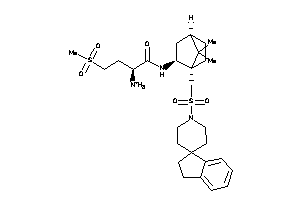
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
54 |
0.27 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.21 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.21 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7400 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.41 |
7.99 |
-59.2 |
4 |
7 |
1 |
111 |
550.811 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.41 |
7.66 |
-24.93 |
3 |
7 |
0 |
110 |
549.803 |
8 |
↓
|
|
|
|
Analogs
-
3915327
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
54 |
0.27 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.21 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.21 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7400 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.41 |
7.97 |
-54.55 |
4 |
7 |
1 |
111 |
550.811 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.41 |
7.64 |
-26.23 |
3 |
7 |
0 |
110 |
549.803 |
8 |
↓
|
|
|
|
Analogs
-
3915327
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OXYR-1-E |
Oxytocin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
54 |
0.27 |
Binding ≤ 10μM
|
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.21 |
Binding ≤ 10μM
|
|
V1BR-1-E |
Vasopressin V1b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.21 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7400 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.41 |
8.08 |
-55.93 |
4 |
7 |
1 |
111 |
550.811 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.41 |
7.75 |
-26.61 |
3 |
7 |
0 |
110 |
549.803 |
8 |
↓
|
|