|
|
Analogs
-
3940300
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
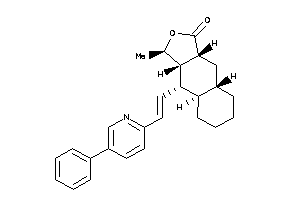
(1R,3aR,4aS,8aR,9S,9aS)-1-methyl-9-[(E)-2-[5-(p-tolyl)-2-pyridyl]vinyl]-3a,4,4a,5,6,7,8,8a,9,9a-deca
(1R,3aR,4aS,8aR,9S,9aS)-1-methyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAR1-1-E |
Proteinase Activated Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
325 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.81 |
15.07 |
-9.84 |
0 |
3 |
0 |
39 |
401.55 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.81 |
15.3 |
-46.02 |
1 |
3 |
1 |
40 |
402.558 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2-[5-(4-methoxyphenyl)-2-pyridyl]vinyl]-3-methyl-3a,4,4a,5,6,7,8,8a,9
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAR1-1-E |
Proteinase Activated Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
204 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.42 |
13.69 |
-11.19 |
0 |
4 |
0 |
48 |
417.549 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.42 |
13.91 |
-48.61 |
1 |
4 |
1 |
50 |
418.557 |
4 |
↓
|
|
|
|
Analogs
-
3955418
-
-
3940301
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
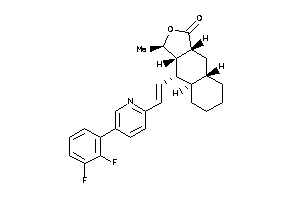
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2-[5-(4-fluorophenyl)-2-pyridyl]vinyl]-3-methyl-3a,4,4a,5,6,7,8,8a,9,
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAR1-1-E |
Proteinase Activated Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
467 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.53 |
14.46 |
-10.57 |
0 |
3 |
0 |
39 |
405.513 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.53 |
14.69 |
-50.86 |
1 |
3 |
1 |
40 |
406.521 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2-[5-(4-chlorophenyl)-2-pyridyl]vinyl]-3-methyl-3a,4,4a,5,6,7,8,8a,9,
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAR1-1-E |
Proteinase Activated Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.04 |
14.91 |
-9.99 |
0 |
3 |
0 |
39 |
421.968 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.04 |
15.14 |
-50.05 |
1 |
3 |
1 |
40 |
422.976 |
3 |
↓
|
|
|
|
Analogs
-
3940302
-
-
3940296
-
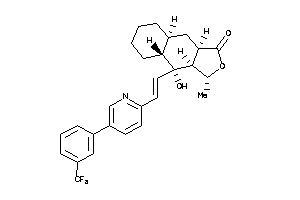
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,3aS,4S,4aR,8aS,9aR)-3-methyl-4-[(E)-2-[5-[4-(trifluoromethyl)phenyl]-2-pyridyl]vinyl]-3a,4,4a,5,
(3R,3aS,4S,4aR,8aS,9aR)-3-methyl…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.26 |
15.39 |
-10.82 |
0 |
3 |
0 |
39 |
455.52 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.26 |
15.63 |
-52.8 |
1 |
3 |
1 |
40 |
456.528 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aR,4aS,8aR,9S,9aS)-1-methyl-9-[(E)-2-[5-(o-tolyl)-2-pyridyl]vinyl]-3a,4,4a,5,6,7,8,8a,9,9a-deca
(1R,3aR,4aS,8aR,9S,9aS)-1-methyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAR1-1-E |
Proteinase Activated Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
14 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.76 |
14.99 |
-9.23 |
0 |
3 |
0 |
39 |
401.55 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.76 |
15.22 |
-45.54 |
1 |
3 |
1 |
40 |
402.558 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,3aS,4S,4aR,8aS,9aR)-3-methyl-4-[(E)-2-[5-[2-(trifluoromethyl)phenyl]-2-pyridyl]vinyl]-3a,4,4a,5,
(3R,3aS,4S,4aR,8aS,9aR)-3-methyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAR1-1-E |
Proteinase Activated Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
46 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.21 |
15.28 |
-10.63 |
0 |
3 |
0 |
39 |
455.52 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.21 |
15.5 |
-45.9 |
1 |
3 |
1 |
40 |
456.528 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAR1-1-E |
Proteinase Activated Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
44 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.52 |
15.97 |
-12.26 |
0 |
5 |
0 |
66 |
459.586 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.52 |
16.18 |
-43.11 |
1 |
5 |
1 |
67 |
460.594 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2-[5-(2-methoxyphenyl)-2-pyridyl]vinyl]-3-methyl-3a,4,4a,5,6,7,8,8a,9
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAR1-1-E |
Proteinase Activated Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
11 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.37 |
14.01 |
-10.93 |
0 |
4 |
0 |
48 |
417.549 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.37 |
14.23 |
-42.56 |
1 |
4 |
1 |
50 |
418.557 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2-[5-(2-chlorophenyl)-2-pyridyl]vinyl]-3-methyl-3a,4,4a,5,6,7,8,8a,9,
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAR1-1-E |
Proteinase Activated Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.99 |
14.91 |
-10.31 |
0 |
3 |
0 |
39 |
421.968 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.99 |
15.13 |
-43.28 |
1 |
3 |
1 |
40 |
422.976 |
3 |
↓
|
|
|
|
Analogs
-
3955418
-

-
3940301
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
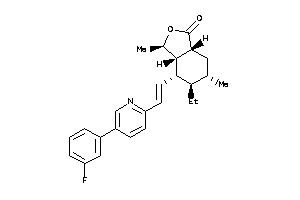
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2-[5-(3-fluorophenyl)-2-pyridyl]vinyl]-3-methyl-3a,4,4a,5,6,7,8,8a,9,
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAR1-1-E |
Proteinase Activated Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
35 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.50 |
14.46 |
-10.51 |
0 |
3 |
0 |
39 |
405.513 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.50 |
14.69 |
-50.26 |
1 |
3 |
1 |
40 |
406.521 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2-[5-(3-bromophenyl)-2-pyridyl]vinyl]-3-methyl-3a,4,4a,5,6,7,8,8a,9,9
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAR1-1-E |
Proteinase Activated Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.15 |
15.02 |
-9.9 |
0 |
3 |
0 |
39 |
466.419 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.15 |
15.24 |
-49.37 |
1 |
3 |
1 |
40 |
467.427 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
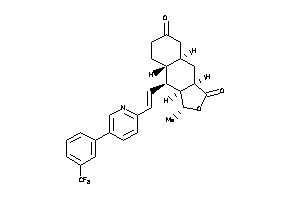
3-[6-[(E)-2-[(3R,3aS,4S,4aR,8aS,9aR)-3-methyl-1-oxo-3a,4,4a,5,6,7,8,8a,9,9a-decahydro-3H-benzo[f]iso
3-[6-[(E)-2-[(3R,3aS,4S,4aR,8aS,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAR1-1-E |
Proteinase Activated Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.09 |
14.78 |
-13.62 |
0 |
4 |
0 |
63 |
412.533 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.09 |
15 |
-53.82 |
1 |
4 |
1 |
64 |
413.541 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aR,4aS,8aR,9S,9aS)-1-methyl-9-[(E)-2-[5-(m-tolyl)-2-pyridyl]vinyl]-3a,4,4a,5,6,7,8,8a,9,9a-deca
(1R,3aR,4aS,8aR,9S,9aS)-1-methyl…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAR1-1-E |
Proteinase Activated Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
13 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.79 |
15.06 |
-9.86 |
0 |
3 |
0 |
39 |
401.55 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.79 |
15.28 |
-46.1 |
1 |
3 |
1 |
40 |
402.558 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2-[5-(3-isopropylphenyl)-2-pyridyl]vinyl]-3-methyl-3a,4,4a,5,6,7,8,8a
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAR1-1-E |
Proteinase Activated Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
19 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.85 |
16.3 |
-9.54 |
0 |
3 |
0 |
39 |
429.604 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.85 |
16.53 |
-46.18 |
1 |
3 |
1 |
40 |
430.612 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2-[5-(3-methoxyphenyl)-2-pyridyl]vinyl]-3-methyl-3a,4,4a,5,6,7,8,8a,9
(3R,3aS,4S,4aR,8aS,9aR)-4-[(E)-2…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAR1-1-E |
Proteinase Activated Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
28 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.39 |
13.69 |
-11.57 |
0 |
4 |
0 |
48 |
417.549 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.39 |
13.91 |
-46.91 |
1 |
4 |
1 |
50 |
418.557 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[6-[(E)-2-[(3R,3aS,4S,4aR,8aS,9aR)-3-methyl-1-oxo-3a,4,4a,5,6,7,8,8a,9,9a-decahydro-3H-benzo[f]iso
3-[6-[(E)-2-[(3R,3aS,4S,4aR,8aS,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAR1-1-E |
Proteinase Activated Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
9.39 |
-17.66 |
2 |
6 |
0 |
99 |
466.603 |
4 |
↓
|
|