|
|
Analogs
-
34043163
-
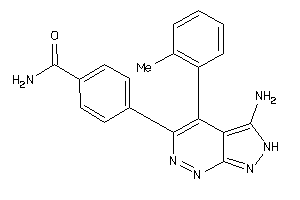
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.00 |
14.09 |
-16.42 |
3 |
5 |
0 |
80 |
439.522 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
7.00 |
14.11 |
-9.69 |
3 |
5 |
0 |
80 |
439.522 |
4 |
↓
|
|
|
|
Analogs
-
34043163
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
7.89 |
-23.77 |
3 |
11 |
0 |
172 |
377.32 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
3.33 |
8.71 |
-13.27 |
3 |
11 |
0 |
172 |
377.32 |
4 |
↓
|
|
|
|
Analogs
-
34043163
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.21 |
9.27 |
-14.37 |
3 |
5 |
0 |
80 |
423.32 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
5.21 |
9.29 |
-7.98 |
3 |
5 |
0 |
80 |
423.32 |
4 |
↓
|
|
|
|
Analogs
-
34043163
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.91 |
13.39 |
-17.11 |
3 |
5 |
0 |
80 |
439.522 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB-1-E |
G2/mitotic-specific Cyclin B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.41 |
Binding ≤ 10μM |
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1500 |
0.41 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
3.91 |
-15.92 |
3 |
7 |
0 |
107 |
267.248 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
1.70 |
3.91 |
-10.01 |
3 |
7 |
0 |
107 |
267.248 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
4.76 |
-16.62 |
3 |
7 |
0 |
106 |
289.302 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
1.12 |
4.78 |
-11.12 |
3 |
7 |
0 |
106 |
289.302 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.12 |
4.97 |
-46.96 |
4 |
7 |
1 |
108 |
290.31 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB-1-E |
G2/mitotic-specific Cyclin B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.47 |
Binding ≤ 10μM |
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4000 |
0.47 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.33 |
4.1 |
-14.32 |
3 |
5 |
0 |
80 |
211.228 |
1 |
↓
|
|
Ref
Reference (pH 7)
|
1.33 |
4.08 |
-7.9 |
3 |
5 |
0 |
80 |
211.228 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB-1-E |
G2/mitotic-specific Cyclin B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5300 |
0.49 |
Binding ≤ 10μM |
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5300 |
0.49 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.83 |
2.06 |
-17.12 |
3 |
6 |
0 |
94 |
201.189 |
1 |
↓
|
|
Ref
Reference (pH 7)
|
0.83 |
2.09 |
-9.56 |
3 |
6 |
0 |
94 |
201.189 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB-1-E |
G2/mitotic-specific Cyclin B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7500 |
0.27 |
Binding ≤ 10μM |
|
CDK1-3-E |
Cyclin-dependent Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7500 |
0.27 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.82 |
7.91 |
-15.26 |
3 |
7 |
0 |
96 |
358.405 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
3.82 |
7.91 |
-14.65 |
3 |
7 |
0 |
96 |
358.405 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
8.41 |
-16.51 |
2 |
6 |
0 |
84 |
329.363 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
3.15 |
8.41 |
-15.08 |
2 |
6 |
0 |
84 |
329.363 |
3 |
↓
|
|