|
|
Analogs
-
26144702
-
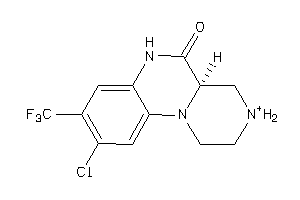
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.56 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
516 |
0.44 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
5-hydroxytryptamine Receptor 2C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
136 |
0.48 |
Functional ≤ 10μM
|
|
5HT2C-2-E |
5-hydroxytryptamine Receptor 2C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
2.55 |
-53.32 |
3 |
4 |
1 |
49 |
306.695 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.79 |
1.13 |
-7.42 |
2 |
4 |
0 |
44 |
305.687 |
1 |
↓
|
|
|
|
Analogs
-
26144695
-
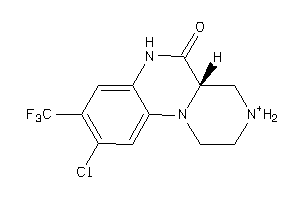
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.56 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.53 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
5-hydroxytryptamine Receptor 2C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
19 |
0.54 |
Functional ≤ 10μM
|
|
5HT2C-2-E |
5-hydroxytryptamine Receptor 2C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
136 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
2.56 |
-53.33 |
3 |
4 |
1 |
49 |
306.695 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.79 |
1.16 |
-6.93 |
2 |
4 |
0 |
44 |
305.687 |
1 |
↓
|
|
|
|
Analogs
-
26150750
-
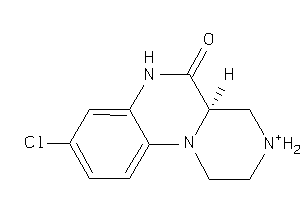
-
26150757
-
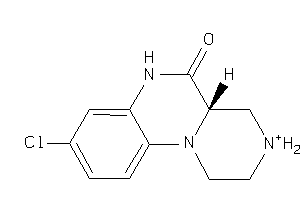
-
29571960
-
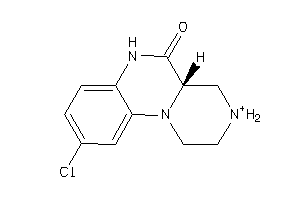
-
3966376
-
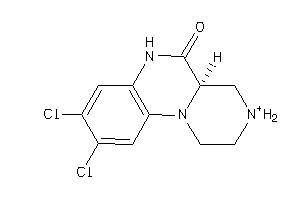
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aR)-9-chloro-1,2,3,4,4a,6-hexahydropyrazino[2,1-c]quinoxalin-5-one
(4aR)-9-chloro-1,2,3,4,4a,6-hexa…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
34 |
0.65 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
5-hydroxytryptamine Receptor 2C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
86 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.97 |
1.67 |
-48.23 |
3 |
4 |
1 |
49 |
238.698 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.97 |
0.28 |
-6.83 |
2 |
4 |
0 |
44 |
237.69 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.16 |
3.81 |
-8.01 |
1 |
5 |
0 |
48 |
312.348 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.16 |
6.09 |
-49.73 |
2 |
5 |
1 |
50 |
313.356 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.16 |
3.8 |
-8.02 |
1 |
5 |
0 |
48 |
312.348 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.16 |
6.09 |
-49.69 |
2 |
5 |
1 |
50 |
313.356 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.21 |
8.42 |
-48.03 |
2 |
5 |
1 |
50 |
337.447 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.21 |
6.16 |
-9.75 |
1 |
5 |
0 |
48 |
336.439 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.21 |
8.88 |
-90.7 |
3 |
5 |
2 |
51 |
338.455 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.21 |
8.43 |
-48.06 |
2 |
5 |
1 |
50 |
337.447 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.21 |
6.16 |
-9.76 |
1 |
5 |
0 |
48 |
336.439 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.21 |
8.89 |
-90.67 |
3 |
5 |
2 |
51 |
338.455 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.72 |
9.2 |
-46.02 |
2 |
5 |
1 |
50 |
351.474 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.72 |
6.94 |
-9.33 |
1 |
5 |
0 |
48 |
350.466 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.72 |
9.66 |
-87.57 |
3 |
5 |
2 |
51 |
352.482 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.72 |
9.21 |
-46.04 |
2 |
5 |
1 |
50 |
351.474 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.72 |
6.94 |
-9.34 |
1 |
5 |
0 |
48 |
350.466 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.72 |
9.67 |
-87.58 |
3 |
5 |
2 |
51 |
352.482 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.26 |
5.5 |
-8.48 |
0 |
5 |
0 |
40 |
308.385 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.26 |
7.78 |
-45.76 |
1 |
5 |
1 |
41 |
309.393 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.26 |
5.96 |
-39.96 |
1 |
5 |
1 |
41 |
309.393 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
5.39 |
-44.09 |
3 |
5 |
1 |
52 |
383.438 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.73 |
7.66 |
-124.72 |
4 |
5 |
2 |
53 |
384.446 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
5.4 |
-43.12 |
3 |
5 |
1 |
52 |
383.438 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.73 |
7.65 |
-124.35 |
4 |
5 |
2 |
53 |
384.446 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.56 |
4.67 |
-9.63 |
1 |
5 |
0 |
48 |
326.375 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.56 |
5.13 |
-39.19 |
2 |
5 |
1 |
50 |
327.383 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.56 |
6.95 |
-54.56 |
2 |
5 |
1 |
50 |
327.383 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.56 |
4.66 |
-9.63 |
1 |
5 |
0 |
48 |
326.375 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.56 |
5.12 |
-39.17 |
2 |
5 |
1 |
50 |
327.383 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.56 |
6.95 |
-54.58 |
2 |
5 |
1 |
50 |
327.383 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
4.6 |
-9.48 |
1 |
5 |
0 |
48 |
308.385 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.43 |
6.87 |
-50.44 |
2 |
5 |
1 |
50 |
309.393 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.43 |
5.06 |
-38.17 |
2 |
5 |
1 |
50 |
309.393 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
4.6 |
-9.47 |
1 |
5 |
0 |
48 |
308.385 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.43 |
6.88 |
-50.45 |
2 |
5 |
1 |
50 |
309.393 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.43 |
5.06 |
-38.18 |
2 |
5 |
1 |
50 |
309.393 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
5.59 |
-9.54 |
1 |
5 |
0 |
48 |
376.382 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.42 |
7.87 |
-48.44 |
2 |
5 |
1 |
50 |
377.39 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
7.43 |
-105.07 |
4 |
5 |
2 |
53 |
384.446 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.44 |
5.15 |
-43.72 |
3 |
5 |
1 |
52 |
383.438 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
7.42 |
-105.33 |
4 |
5 |
2 |
53 |
384.446 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.44 |
5.16 |
-44.27 |
3 |
5 |
1 |
52 |
383.438 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.59 |
9.2 |
-43.04 |
2 |
5 |
1 |
50 |
351.474 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.59 |
6.94 |
-9.17 |
1 |
5 |
0 |
48 |
350.466 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.59 |
9.54 |
-88.65 |
3 |
5 |
2 |
51 |
352.482 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.59 |
9.21 |
-43.22 |
2 |
5 |
1 |
50 |
351.474 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.59 |
6.94 |
-9.33 |
1 |
5 |
0 |
48 |
350.466 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.59 |
9.55 |
-88.7 |
3 |
5 |
2 |
51 |
352.482 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.71 |
4.21 |
-43.27 |
3 |
5 |
1 |
52 |
333.431 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.71 |
6.49 |
-103.33 |
4 |
5 |
2 |
53 |
334.439 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.71 |
4.2 |
-43.89 |
3 |
5 |
1 |
52 |
333.431 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.71 |
6.49 |
-103.65 |
4 |
5 |
2 |
53 |
334.439 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
4.14 |
-42.64 |
3 |
5 |
1 |
52 |
315.441 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.57 |
6.41 |
-98.91 |
4 |
5 |
2 |
53 |
316.449 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
4.14 |
-43.19 |
3 |
5 |
1 |
52 |
315.441 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.57 |
6.42 |
-99.25 |
4 |
5 |
2 |
53 |
316.449 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.86 |
4.38 |
-42.12 |
3 |
5 |
1 |
52 |
315.441 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.86 |
6.64 |
-117.76 |
4 |
5 |
2 |
53 |
316.449 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.86 |
4.38 |
-41.24 |
3 |
5 |
1 |
52 |
315.441 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.86 |
6.65 |
-117.44 |
4 |
5 |
2 |
53 |
316.449 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.68 |
5.58 |
-8.62 |
1 |
5 |
0 |
48 |
377.275 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.68 |
7.85 |
-54.77 |
2 |
5 |
1 |
50 |
378.283 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.68 |
6.04 |
-38.28 |
2 |
5 |
1 |
50 |
378.283 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
8.5 |
-51.89 |
2 |
5 |
1 |
50 |
355.437 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.35 |
6.23 |
-9.84 |
1 |
5 |
0 |
48 |
354.429 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.35 |
8.96 |
-94.99 |
3 |
5 |
2 |
51 |
356.445 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
8.5 |
-51.89 |
2 |
5 |
1 |
50 |
355.437 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.35 |
6.22 |
-9.86 |
1 |
5 |
0 |
48 |
354.429 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.35 |
8.96 |
-95.01 |
3 |
5 |
2 |
51 |
356.445 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.54 |
4.59 |
-10.01 |
1 |
5 |
0 |
48 |
308.385 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.54 |
6.85 |
-43.39 |
2 |
5 |
1 |
50 |
309.393 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.54 |
4.58 |
-9.37 |
1 |
5 |
0 |
48 |
308.385 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.54 |
6.86 |
-43.17 |
2 |
5 |
1 |
50 |
309.393 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.94 |
5.38 |
-9.2 |
1 |
5 |
0 |
48 |
322.412 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.94 |
7.64 |
-48.07 |
2 |
5 |
1 |
50 |
323.42 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.94 |
8.1 |
-96.53 |
3 |
5 |
2 |
51 |
324.428 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.94 |
5.38 |
-9.2 |
1 |
5 |
0 |
48 |
322.412 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.94 |
7.65 |
-48.14 |
2 |
5 |
1 |
50 |
323.42 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.94 |
8.11 |
-96.6 |
3 |
5 |
2 |
51 |
324.428 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.02 |
3.74 |
-8.05 |
1 |
5 |
0 |
48 |
294.358 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.02 |
6.02 |
-45.81 |
2 |
5 |
1 |
50 |
295.366 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.02 |
3.73 |
-8.01 |
1 |
5 |
0 |
48 |
294.358 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.02 |
6.02 |
-45.83 |
2 |
5 |
1 |
50 |
295.366 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
5.61 |
-9.94 |
1 |
5 |
0 |
48 |
376.382 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.30 |
7.89 |
-56.1 |
2 |
5 |
1 |
50 |
377.39 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.30 |
6.07 |
-39.6 |
2 |
5 |
1 |
50 |
377.39 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
5.61 |
-9.97 |
1 |
5 |
0 |
48 |
376.382 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.30 |
6.07 |
-39.58 |
2 |
5 |
1 |
50 |
377.39 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.30 |
7.88 |
-56.1 |
2 |
5 |
1 |
50 |
377.39 |
4 |
↓
|
|