|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.48 |
4.4 |
-36.91 |
0 |
4 |
-1 |
53 |
191.189 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.48 |
4.4 |
-6.91 |
1 |
4 |
0 |
54 |
192.197 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.10 |
4.81 |
-38.54 |
0 |
4 |
-1 |
53 |
252.095 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.10 |
4.81 |
-5.73 |
1 |
4 |
0 |
54 |
253.103 |
3 |
↓
|
|
|
|
Analogs
-
36143328
-
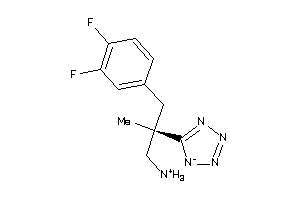
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CA2D1-3-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
520 |
0.49 |
Binding ≤ 10μM
|
|
CA2D2-2-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.49 |
Binding ≤ 10μM
|
|
CA2D3-2-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.49 |
Binding ≤ 10μM
|
|
CA2D4-2-E |
Voltage-gated Calcium Channel Alpha2/delta-4 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.82 |
2.17 |
-40.58 |
3 |
5 |
0 |
80 |
253.256 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.82 |
2.33 |
-58.32 |
4 |
5 |
1 |
82 |
254.264 |
4 |
↓
|
|
|
|
Analogs
-
36143325
-
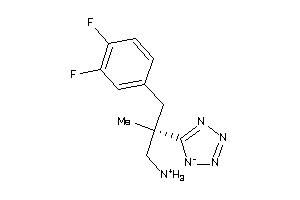
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CA2D1-3-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
520 |
0.49 |
Binding ≤ 10μM
|
|
CA2D2-2-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.49 |
Binding ≤ 10μM
|
|
CA2D3-2-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.49 |
Binding ≤ 10μM
|
|
CA2D4-2-E |
Voltage-gated Calcium Channel Alpha2/delta-4 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.82 |
2.19 |
-46.35 |
3 |
5 |
0 |
80 |
253.256 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.82 |
2.35 |
-59.43 |
4 |
5 |
1 |
82 |
254.264 |
4 |
↓
|
|
|
|
Analogs
-
36143335
-
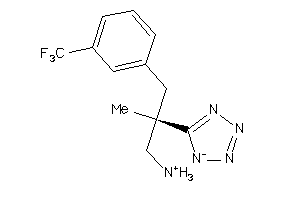
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CA2D1-3-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
160 |
0.48 |
Binding ≤ 10μM
|
|
CA2D2-2-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.48 |
Binding ≤ 10μM
|
|
CA2D3-2-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.48 |
Binding ≤ 10μM
|
|
CA2D4-2-E |
Voltage-gated Calcium Channel Alpha2/delta-4 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
3.09 |
-37.78 |
3 |
5 |
0 |
80 |
285.273 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.43 |
3.25 |
-53.61 |
4 |
5 |
1 |
82 |
286.281 |
5 |
↓
|
|
|
|
Analogs
-
36143331
-
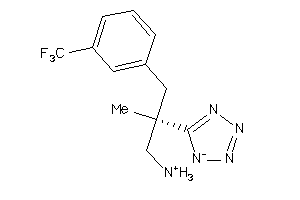
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CA2D1-3-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
160 |
0.48 |
Binding ≤ 10μM
|
|
CA2D2-2-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.48 |
Binding ≤ 10μM
|
|
CA2D3-2-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.48 |
Binding ≤ 10μM
|
|
CA2D4-2-E |
Voltage-gated Calcium Channel Alpha2/delta-4 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
3.09 |
-45.06 |
3 |
5 |
0 |
80 |
285.273 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.43 |
3.25 |
-56.3 |
4 |
5 |
1 |
82 |
286.281 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.65 |
6.69 |
-42.64 |
0 |
6 |
-1 |
84 |
231.235 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.10 |
7.36 |
-42.7 |
0 |
6 |
-1 |
84 |
245.262 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.92 |
7.24 |
-39.92 |
0 |
6 |
-1 |
84 |
245.262 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
7.92 |
-40.04 |
0 |
6 |
-1 |
84 |
259.289 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.19 |
8.04 |
-48.28 |
0 |
6 |
-1 |
84 |
259.289 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.64 |
8.72 |
-48.33 |
0 |
6 |
-1 |
84 |
273.316 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.10 |
8.55 |
-14.06 |
0 |
6 |
0 |
64 |
315.421 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
6.68 |
-15.14 |
0 |
6 |
0 |
70 |
246.27 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.74 |
4.81 |
-37.46 |
0 |
4 |
-1 |
53 |
187.226 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.74 |
4.95 |
-8.93 |
1 |
4 |
0 |
54 |
188.234 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.76 |
5 |
-37.58 |
0 |
4 |
-1 |
53 |
187.226 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.76 |
5.14 |
-8.99 |
1 |
4 |
0 |
54 |
188.234 |
3 |
↓
|
|
|
|
Analogs
-
41211181
-
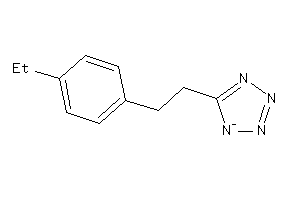
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
5.01 |
-37.46 |
0 |
4 |
-1 |
53 |
187.226 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.79 |
5.15 |
-8.95 |
1 |
4 |
0 |
54 |
188.234 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.46 |
4.37 |
-40 |
0 |
4 |
-1 |
53 |
191.189 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.46 |
4.5 |
-8.7 |
1 |
4 |
0 |
54 |
192.197 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.50 |
4.4 |
-35.66 |
0 |
4 |
-1 |
53 |
191.189 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.50 |
4.54 |
-10.54 |
1 |
4 |
0 |
54 |
192.197 |
3 |
↓
|
|
|
|
Analogs
-
41204169
-
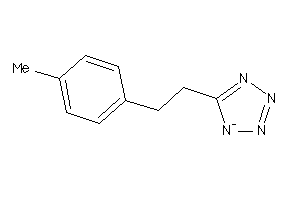
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.25 |
5.77 |
-37.28 |
0 |
4 |
-1 |
53 |
201.253 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.25 |
5.91 |
-8.79 |
1 |
4 |
0 |
54 |
202.261 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.35 |
3.69 |
-40.93 |
0 |
5 |
-1 |
62 |
203.225 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.35 |
3.83 |
-8.95 |
1 |
5 |
0 |
64 |
204.233 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
3.63 |
-39.15 |
0 |
5 |
-1 |
62 |
203.225 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.37 |
3.77 |
-10.88 |
1 |
5 |
0 |
64 |
204.233 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
3.63 |
-37.78 |
0 |
5 |
-1 |
62 |
203.225 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.40 |
3.77 |
-10.77 |
1 |
5 |
0 |
64 |
204.233 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
4.74 |
-38.74 |
0 |
4 |
-1 |
53 |
207.644 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.97 |
4.88 |
-8 |
1 |
4 |
0 |
54 |
208.652 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.99 |
4.84 |
-36.48 |
0 |
4 |
-1 |
53 |
207.644 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.99 |
4.98 |
-10.13 |
1 |
4 |
0 |
54 |
208.652 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
4.85 |
-35.37 |
0 |
4 |
-1 |
53 |
207.644 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.02 |
4.99 |
-9.73 |
1 |
4 |
0 |
54 |
208.652 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
6.26 |
-37.1 |
0 |
4 |
-1 |
53 |
215.28 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.85 |
6.4 |
-8.71 |
1 |
4 |
0 |
54 |
216.288 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.44 |
4.74 |
-38.79 |
0 |
5 |
-1 |
56 |
216.268 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.72 |
4.62 |
-41.18 |
0 |
5 |
-1 |
62 |
217.252 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.72 |
4.75 |
-8.7 |
1 |
5 |
0 |
64 |
218.26 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
4.56 |
-39.05 |
0 |
5 |
-1 |
62 |
217.252 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.75 |
4.7 |
-10.77 |
1 |
5 |
0 |
64 |
218.26 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.77 |
4.56 |
-37.59 |
0 |
5 |
-1 |
62 |
217.252 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.77 |
4.7 |
-10.67 |
1 |
5 |
0 |
64 |
218.26 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
4.78 |
-40.69 |
0 |
4 |
-1 |
53 |
225.634 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.08 |
4.92 |
-8.86 |
1 |
4 |
0 |
54 |
226.642 |
3 |
↓
|
|