|
|
Analogs
-
43968061
-
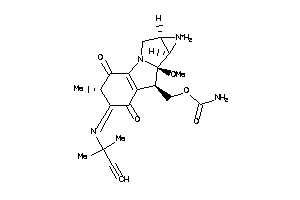
-
43968063
-
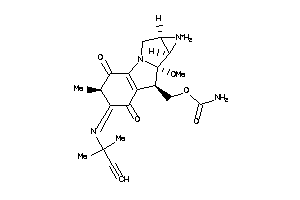
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.82 |
-2.41 |
-14.77 |
3 |
9 |
0 |
133 |
372.381 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.82 |
-2.44 |
-16.86 |
3 |
9 |
0 |
133 |
372.381 |
5 |
↓
|
|
|
|
Analogs
-
43968061
-

-
43968063
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.82 |
-3.18 |
-13.91 |
3 |
9 |
0 |
133 |
372.381 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.82 |
-3.23 |
-15.68 |
3 |
9 |
0 |
133 |
372.381 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.72 |
0.73 |
-15.19 |
3 |
9 |
0 |
133 |
410.43 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.72 |
1.49 |
-57.95 |
4 |
9 |
1 |
128 |
411.438 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.72 |
0 |
-14.34 |
3 |
9 |
0 |
133 |
410.43 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.72 |
1.53 |
-49.3 |
4 |
9 |
1 |
128 |
411.438 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
0.76 |
-12.77 |
3 |
9 |
0 |
133 |
444.875 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.39 |
1.6 |
-56.31 |
4 |
9 |
1 |
128 |
445.883 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.39 |
0.93 |
-12.83 |
3 |
9 |
0 |
133 |
444.875 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
0.28 |
-11.98 |
3 |
9 |
0 |
133 |
444.875 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.39 |
1.88 |
-51.31 |
4 |
9 |
1 |
128 |
445.883 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.39 |
0.34 |
-12.23 |
3 |
9 |
0 |
133 |
444.875 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.88 |
0.3 |
-13.08 |
3 |
9 |
0 |
133 |
428.42 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.88 |
1.14 |
-56.72 |
4 |
9 |
1 |
128 |
429.428 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.88 |
0.48 |
-13.13 |
3 |
9 |
0 |
133 |
428.42 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.88 |
-0.17 |
-12.35 |
3 |
9 |
0 |
133 |
428.42 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.88 |
1.44 |
-51.65 |
4 |
9 |
1 |
128 |
429.428 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.88 |
-0.11 |
-12.54 |
3 |
9 |
0 |
133 |
428.42 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.45 |
1.14 |
-15.13 |
3 |
10 |
0 |
157 |
435.44 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.45 |
1.87 |
-59.33 |
4 |
10 |
1 |
152 |
436.448 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.45 |
0.39 |
-14.66 |
3 |
10 |
0 |
157 |
435.44 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.45 |
1.92 |
-53.41 |
4 |
10 |
1 |
152 |
436.448 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
5500 |
0.23 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81247 |
Z81247
|
HeLa (Cervical Adenocarcinoma Cells) |
5500 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.02 |
-1.72 |
-13.96 |
4 |
10 |
0 |
154 |
440.456 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.02 |
-1.72 |
-14.78 |
4 |
10 |
0 |
154 |
440.456 |
8 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
1000 |
0.27 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81247 |
Z81247
|
HeLa (Cervical Adenocarcinoma Cells) |
1000 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.46 |
-0.17 |
-13.25 |
4 |
9 |
0 |
145 |
442.497 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.46 |
-0.18 |
-12.19 |
4 |
9 |
0 |
145 |
442.497 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
2200 |
0.25 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81247 |
Z81247
|
HeLa (Cervical Adenocarcinoma Cells) |
2200 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.52 |
0.27 |
-13.51 |
4 |
9 |
0 |
145 |
456.524 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.52 |
0.28 |
-14.43 |
4 |
9 |
0 |
145 |
456.524 |
8 |
↓
|
|
|
|
Analogs
-
43693386
-
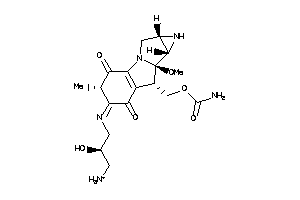
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.10 |
-2.6 |
-11.25 |
3 |
9 |
0 |
133 |
376.413 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.35 |
-8.44 |
-17.79 |
6 |
11 |
0 |
185 |
440.478 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.35 |
-7.02 |
-52.34 |
7 |
11 |
0 |
180 |
441.486 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.35 |
-8.44 |
-18.64 |
6 |
11 |
0 |
185 |
440.478 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
1.83 |
-21.63 |
3 |
10 |
0 |
143 |
470.507 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.94 |
1.83 |
-23.39 |
3 |
10 |
0 |
143 |
470.507 |
6 |
↓
|
|
|
|
Analogs
-
14243039
-
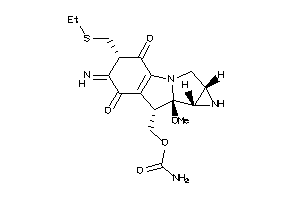
-
14243044
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
1100 |
0.29 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81247 |
Z81247
|
HeLa (Cervical Adenocarcinoma Cells) |
1100 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.21 |
-1.29 |
-12.08 |
4 |
9 |
0 |
145 |
420.491 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.21 |
0.13 |
-47.89 |
5 |
9 |
0 |
139 |
421.499 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.21 |
-1.29 |
-12.91 |
4 |
9 |
0 |
145 |
420.491 |
8 |
↓
|
|