|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1R,3aS,5aR,5bR,7aR,9S,11aR,11bR,13aR,13bR)-3a-(acetoxymethyl)-1-isopropenyl-5a,5b,8,8,11a-pentamet
[(1R,3aS,5aR,5bR,7aR,9S,11aR,11b…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.11 |
18.55 |
-9.07 |
0 |
6 |
0 |
70 |
592.865 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.11 |
18.96 |
-34.32 |
1 |
6 |
1 |
72 |
593.873 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1R,3aS,5aR,5bR,7aR,9S,11aR,11bR,13aR,13bS)-3a-methoxycarbonyl-1-[1-(methoxymethyl)vinyl]-5a,5b,8,8
[(1R,3aS,5aR,5bR,7aR,9S,11aR,11b…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.42 |
17.4 |
-9.73 |
0 |
7 |
0 |
80 |
608.864 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.42 |
17.91 |
-39.14 |
1 |
7 |
1 |
81 |
609.872 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1R,3aS,5aR,5bR,7aR,9S,11aR,11bR,13aR,13bR)-9-hydroxy-5a,5b,8,8,11a-pentamethyl-1-[(1R)-1-methyl-2-
[(1R,3aS,5aR,5bR,7aR,9S,11aR,11b…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.79 |
13.58 |
-9.09 |
1 |
6 |
0 |
81 |
566.827 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.79 |
14.03 |
-38.03 |
2 |
6 |
1 |
83 |
567.835 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1R,3aS,5aR,5bR,7aR,9S,11aR,11bR,13aR,13bS)-9-hydroxy-5a,5b,8,8,11a-pentamethyl-1-[(1R)-1-methyl-2-
[(1R,3aS,5aR,5bR,7aR,9S,11aR,11b…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.79 |
13.67 |
-9.03 |
1 |
6 |
0 |
81 |
566.827 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.79 |
14.13 |
-35.82 |
2 |
6 |
1 |
83 |
567.835 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1R,3aS,5aR,5bR,7aR,9S,11aR,11bR,13aR,13bR)-9-hydroxy-1-isopropenyl-5a,5b,8,8,11a-pentamethyl-1,2,3
[(1R,3aS,5aR,5bR,7aR,9S,11aR,11b…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.42 |
14.5 |
-6.27 |
1 |
5 |
0 |
64 |
550.828 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.42 |
14.96 |
-33 |
2 |
5 |
1 |
66 |
551.836 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1R,3aS,5aR,5bR,7aR,9S,11aR,11bR,13aR,13bR)-9-hydroxy-1-isopropenyl-5a,5b,8,8,11a-pentamethyl-1,2,3
[(1R,3aS,5aR,5bR,7aR,9S,11aR,11b…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.12 |
16.79 |
-7.39 |
1 |
4 |
0 |
56 |
602.9 |
7 |
↓
|
|
|
|
Analogs
-
3978802
-
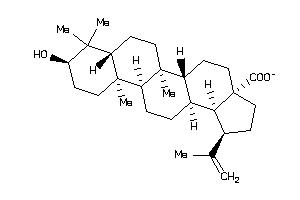
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1S,3aR,5aS,5bS,7aS,9R,11aS,11bS,13aS,13bR)-9-hydroxy-1-isopropenyl-5a,5b,8,8,11a-pentamethyl-1,2,3,
(1S,3aR,5aS,5bS,7aS,9R,11aS,11bS…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.04 |
11.79 |
-56.95 |
1 |
3 |
-1 |
60 |
455.703 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.04 |
9.87 |
-6.46 |
2 |
3 |
0 |
58 |
456.711 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.52 |
7.97 |
-7.67 |
2 |
3 |
0 |
58 |
444.7 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,5aR,5bR,7aR,9S,11aR,11bR,13aR,13bR)-1-isopropenyl-5a,5b,8,8,11a-pentamethyl-9-propanoyloxy-1
(1R,3aS,5aR,5bR,7aR,9S,11aR,11bR…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.10 |
17.04 |
-53.33 |
0 |
4 |
-1 |
66 |
511.767 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.10 |
15.11 |
-8.02 |
1 |
4 |
0 |
64 |
512.775 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,5aR,5bR,7aR,9S,11aR,11bR,13aR,13bS)-3a-(hydroxymethyl)-1-[1-[[2-(4-hydroxyphenyl)ethylamino]
(1R,3aS,5aR,5bR,7aR,9S,11aR,11bR…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.31 |
10.31 |
-52.52 |
5 |
4 |
1 |
77 |
578.902 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
2-[2-[(1R,3aS,5aR,5bR,7aR,9R,11aR,11bR,13aR,13bS)-3a-(hydroxymethyl)-9-(2-methoxyethylamino)-5a,5b,8
2-[2-[(1R,3aS,5aR,5bR,7aR,9R,11a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.76 |
8.47 |
-89.11 |
6 |
5 |
2 |
83 |
560.908 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.76 |
7.57 |
-40.56 |
5 |
5 |
1 |
78 |
559.9 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
4-[2-[[(1R,3aS,5aR,5bR,7aR,9R,11aR,11bR,13aR,13bR)-3a-(hydroxymethyl)-1-isopropenyl-5a,5b,8,8,11a-pe
4-[2-[[(1R,3aS,5aR,5bR,7aR,9R,11…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.35 |
11.19 |
-51.81 |
5 |
4 |
1 |
77 |
578.902 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,5aR,5bR,7aR,9R,11aS,11bR,12R,13aR,13bR)-9,12-dihydroxy-1-isopropenyl-5a,5b,8,8,11a-pentameth
(1R,3aS,5aR,5bR,7aR,9R,11aS,11bR…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.13 |
9.25 |
-46.05 |
2 |
4 |
-1 |
81 |
471.702 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.13 |
7.58 |
-6.2 |
3 |
4 |
0 |
78 |
472.71 |
2 |
↓
|
|