|
|
Analogs
-
4131708
-
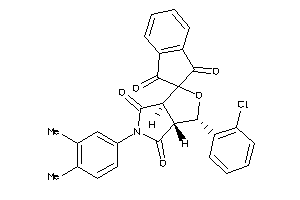
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.71 |
13.95 |
-15.1 |
0 |
6 |
0 |
81 |
485.923 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
4.71 |
12.53 |
-11.22 |
0 |
6 |
0 |
81 |
485.923 |
2 |
↓
|
|
|
|
Analogs
-
3974230
-
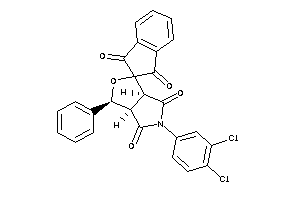
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SYFM-1-E |
Phenylalanyl-tRNA Synthetase Mitochondrial (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.22 |
Binding ≤ 10μM
|
|
SYFA-1-B |
Phenylalanyl-tRNA Synthetase Alpha Chain (cluster #1 Of 1), Bacterial |
Bacteria |
5 |
0.34 |
Binding ≤ 10μM
|
|
SYFA-1-B |
Phenylalanyl-tRNA Synthetase Alpha Chain (cluster #1 Of 1), Bacterial |
Bacteria |
850 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.54 |
11.81 |
-11.12 |
0 |
6 |
0 |
81 |
492.314 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
4.54 |
13.11 |
-15.81 |
0 |
6 |
0 |
81 |
492.314 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
4.54 |
11.72 |
-9.09 |
0 |
6 |
0 |
81 |
492.314 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.34 |
12.9 |
-17.69 |
0 |
7 |
0 |
90 |
501.922 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
4.34 |
11.51 |
-13.8 |
0 |
7 |
0 |
90 |
501.922 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.68 |
13.05 |
-12.71 |
0 |
6 |
0 |
81 |
485.923 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.45 |
12.14 |
-11.11 |
0 |
6 |
0 |
81 |
489.886 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
11.56 |
-15.36 |
0 |
8 |
0 |
99 |
497.503 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
3.74 |
9.99 |
-15.09 |
0 |
8 |
0 |
99 |
497.503 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
11.69 |
-19.63 |
0 |
8 |
0 |
99 |
497.503 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
3.74 |
10.29 |
-14.89 |
0 |
8 |
0 |
99 |
497.503 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.84 |
12.59 |
-17.5 |
0 |
9 |
0 |
127 |
502.866 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
N-[4-[(3S,3aS,6aR)-1',3',4,6-tetraoxo-3-(p-tolyl)spiro[3a,6a-dihydro-3H-furo[3,4-c]pyrrole-1,2'-inda
N-[4-[(3S,3aS,6aR)-1',3',4,6-tet…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
10.01 |
-17 |
1 |
8 |
0 |
110 |
494.503 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
N-[4-[(3R,3aS,6aR)-1',3',4,6-tetraoxo-3-(p-tolyl)spiro[3a,6a-dihydro-3H-furo[3,4-c]pyrrole-1,2'-inda
N-[4-[(3R,3aS,6aR)-1',3',4,6-tet…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
10.32 |
-16.53 |
1 |
8 |
0 |
110 |
494.503 |
3 |
↓
|
|