|
|
Analogs
-
119967
-
-
155356
-
-
525571
-
-
1069133
-
-
3881689
-
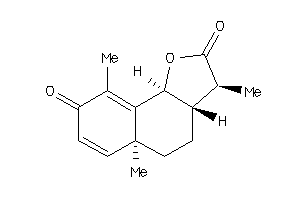
Draw
Identity
99%
90%
80%
70%
Popular Name:
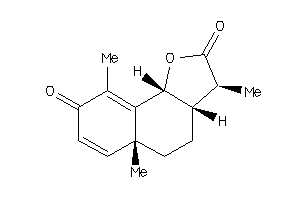
3,5a,9-trimethyl-3a,5,5a,9b-tetrahydronaphtho[1,2-b]furan-2,8(3H,4H)-dione
3,5a,9-trimethyl-3a,5,5a,9b-tetr…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.31 |
7.84 |
-13.76 |
0 |
3 |
0 |
43 |
246.306 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
57495
-
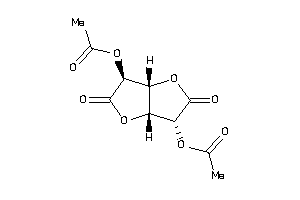
-
3871366
-
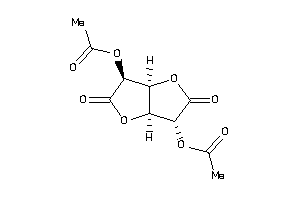
-
3871367
-
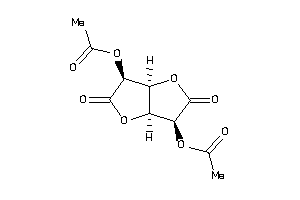
-
11616549
-
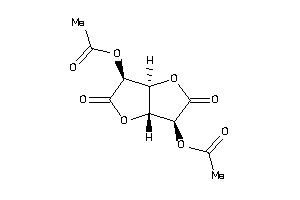
Draw
Identity
99%
90%
80%
70%
Popular Name:
2,5-Di-O-acetyl-D-glucaro-1,4:6,3-dilactone; 2,5-Di-O-acetyl-D-glucosaccharo-1,4:6,3-dilactone; Aceglaton; Aceglatona [INN-Spanish]; Aceglatone; Aceglatone [INN]; Aceglatonum [INN-Latin]; C14H18O8; CCRIS 2774; D-Glucaric acid 1,4:6,3-dilactone diacetate
2,5-Di-O-acetyl-D-glucaro-1,4:6,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.95 |
2.6 |
-13.88 |
0 |
8 |
0 |
105 |
258.182 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.82 |
-1.24 |
-18.28 |
2 |
7 |
0 |
93 |
320.345 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.82 |
-1.17 |
-18.55 |
2 |
7 |
0 |
93 |
320.345 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.58 |
8.68 |
-17.46 |
0 |
6 |
0 |
70 |
306.705 |
4 |
↓
|
|
|
|
Analogs
-
5778462
-
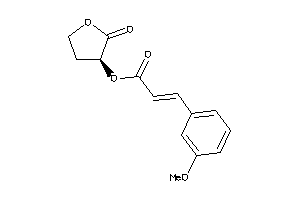
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
2.74 |
-14.77 |
0 |
5 |
0 |
61 |
262.261 |
5 |
↓
|
|
|
|
Analogs
-
5778458
-
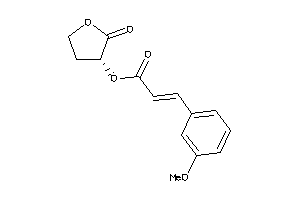
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
2.81 |
-15.17 |
0 |
5 |
0 |
61 |
262.261 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.43 |
4.01 |
-23.63 |
3 |
5 |
0 |
81 |
244.316 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.16 |
0.38 |
-30.5 |
1 |
7 |
0 |
80 |
390.411 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.16 |
0.24 |
-24.73 |
1 |
7 |
0 |
80 |
390.411 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.16 |
8.78 |
-26.91 |
1 |
7 |
0 |
81 |
390.411 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.16 |
0.31 |
-17.57 |
1 |
7 |
0 |
80 |
390.411 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLRA1-1-E |
Glycine Receptor Subunit Alpha-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7100 |
0.24 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLRA1_HUMAN |
P23415
|
Glycine Receptor Subunit Alpha-1, Human |
4200 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.38 |
1.46 |
-21.28 |
3 |
10 |
0 |
149 |
424.402 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLRA1-1-E |
Glycine Receptor Subunit Alpha-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7100 |
0.24 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLRA1_HUMAN |
P23415
|
Glycine Receptor Subunit Alpha-1, Human |
4200 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.38 |
1.96 |
-21.25 |
3 |
10 |
0 |
149 |
424.402 |
1 |
↓
|
|
|
|
Analogs
-
7149678
-
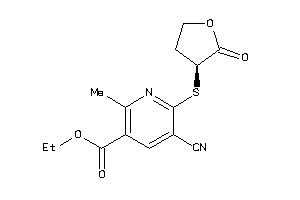
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.75 |
3.22 |
-11.12 |
0 |
6 |
0 |
89 |
306.343 |
5 |
↓
|
|
|
|
Analogs
-
7149674
-
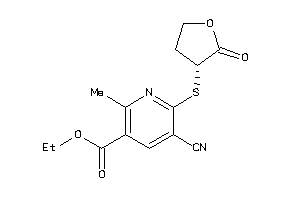
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.75 |
3.22 |
-10.99 |
0 |
6 |
0 |
89 |
306.343 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUOX2-1-E |
Thyroid Oxidase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.28 |
Binding ≤ 10μM
|
|
Z50591-1-O |
Bos Taurus (cluster #1 Of 3), Other |
Other |
3 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.63 |
12.17 |
-13.87 |
4 |
8 |
0 |
126 |
640.943 |
25 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUOX2-1-E |
Thyroid Oxidase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z50591-1-O |
Bos Taurus (cluster #1 Of 3), Other |
Other |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.23 |
16.38 |
-12.01 |
3 |
7 |
0 |
105 |
624.944 |
25 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUOX2-1-E |
Thyroid Oxidase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.27 |
Binding ≤ 10μM
|
|
Z50591-1-O |
Bos Taurus (cluster #1 Of 3), Other |
Other |
1 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.90 |
12.19 |
-13.51 |
4 |
9 |
0 |
138 |
653.942 |
25 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.06 |
5.31 |
-22.07 |
2 |
9 |
0 |
136 |
502.56 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.73 |
5.99 |
-47.35 |
1 |
5 |
1 |
57 |
216.257 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.73 |
3.58 |
-10.99 |
0 |
5 |
0 |
56 |
215.249 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.73 |
5.98 |
-44.27 |
1 |
5 |
1 |
57 |
216.257 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.73 |
3.71 |
-11.03 |
0 |
5 |
0 |
56 |
215.249 |
6 |
↓
|
|
|
|
Analogs
-
33816930
-
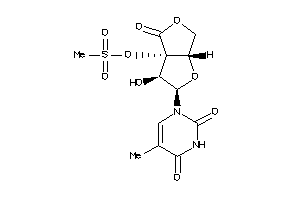
-
40741629
-
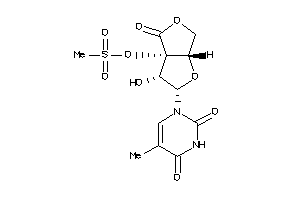
-
40741630
-
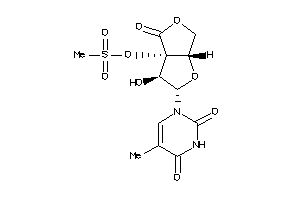
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(2R,3R,3aS,6aS)-3-hydroxy-2-(5-methyl-2,4-dioxo-pyrimidin-1-yl)-4-oxo-2,3,6,6a-tetrahydrofuro[2,3-c
[(2R,3R,3aS,6aS)-3-hydroxy-2-(5-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.08 |
-2.37 |
-23.12 |
2 |
11 |
0 |
154 |
362.316 |
3 |
↓
|
|
|
|
Analogs
-
40741629
-

-
40741630
-

-
33816929
-
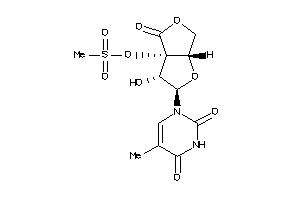
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(2R,3S,3aS,6aS)-3-hydroxy-2-(5-methyl-2,4-dioxo-pyrimidin-1-yl)-4-oxo-2,3,6,6a-tetrahydrofuro[2,3-c
[(2R,3S,3aS,6aS)-3-hydroxy-2-(5-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.08 |
-3.22 |
-20.62 |
2 |
11 |
0 |
154 |
362.316 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.62 |
1.78 |
-39.99 |
5 |
6 |
1 |
100 |
264.305 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.62 |
-0.42 |
-14.67 |
4 |
6 |
0 |
99 |
263.297 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.62 |
1.49 |
-39.2 |
5 |
6 |
1 |
100 |
264.305 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.62 |
-0.38 |
-14.83 |
4 |
6 |
0 |
99 |
263.297 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.64 |
1.78 |
-40.24 |
5 |
6 |
1 |
100 |
264.305 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.64 |
-0.43 |
-14.81 |
4 |
6 |
0 |
99 |
263.297 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.64 |
1.79 |
-36.74 |
5 |
6 |
1 |
100 |
264.305 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.64 |
-0.13 |
-12.18 |
4 |
6 |
0 |
99 |
263.297 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.26 |
2.11 |
-35.56 |
5 |
6 |
1 |
100 |
264.305 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.26 |
-0.27 |
-12.45 |
4 |
6 |
0 |
99 |
263.297 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.26 |
2.93 |
-51.65 |
5 |
6 |
1 |
100 |
264.305 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.26 |
-0.02 |
-14.63 |
4 |
6 |
0 |
99 |
263.297 |
5 |
↓
|
|
|
|
Analogs
-
38436748
-
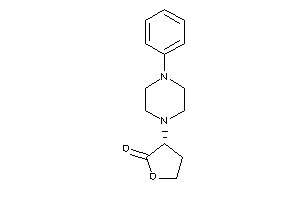
-
38436749
-
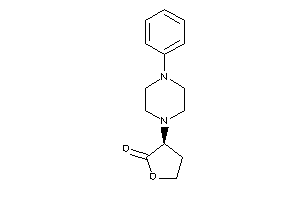
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.18 |
3.39 |
-10.11 |
2 |
5 |
0 |
59 |
261.325 |
2 |
↓
|
|
|
|
Analogs
-
38436748
-

-
38436749
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.18 |
3.37 |
-10.22 |
2 |
5 |
0 |
59 |
261.325 |
2 |
↓
|
|
|
|
Analogs
-
38436748
-

-
38436749
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.20 |
4.32 |
-8.7 |
2 |
5 |
0 |
59 |
261.325 |
2 |
↓
|
|
|
|
Analogs
-
38436748
-

-
38436749
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.20 |
4.3 |
-8.76 |
2 |
5 |
0 |
59 |
261.325 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.48 |
2.94 |
-35.77 |
3 |
6 |
1 |
73 |
263.321 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.48 |
2.46 |
-12.06 |
2 |
6 |
0 |
72 |
262.313 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.48 |
2.92 |
-34.85 |
3 |
6 |
1 |
73 |
263.321 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.48 |
2.45 |
-11.17 |
2 |
6 |
0 |
72 |
262.313 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.48 |
3.2 |
-33.2 |
3 |
6 |
1 |
73 |
263.321 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.48 |
2.88 |
-12.81 |
2 |
6 |
0 |
72 |
262.313 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.48 |
5.35 |
-94.86 |
4 |
6 |
2 |
74 |
264.329 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.48 |
3.16 |
-33.91 |
3 |
6 |
1 |
73 |
263.321 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.48 |
2.85 |
-12.83 |
2 |
6 |
0 |
72 |
262.313 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.48 |
5.34 |
-95.43 |
4 |
6 |
2 |
74 |
264.329 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.70 |
3.37 |
-30.88 |
3 |
6 |
1 |
73 |
263.321 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.70 |
2.9 |
-14.71 |
2 |
6 |
0 |
72 |
262.313 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.70 |
5.68 |
-94.41 |
4 |
6 |
2 |
74 |
264.329 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.70 |
3.51 |
-34.3 |
3 |
6 |
1 |
73 |
263.321 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.70 |
3.04 |
-11.04 |
2 |
6 |
0 |
72 |
262.313 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.70 |
5.68 |
-94.3 |
4 |
6 |
2 |
74 |
264.329 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.89 |
2.93 |
-37.15 |
3 |
6 |
1 |
73 |
263.321 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.89 |
5.08 |
-91.01 |
4 |
6 |
2 |
74 |
264.329 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.89 |
2.91 |
-37.33 |
3 |
6 |
1 |
73 |
263.321 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.89 |
5.08 |
-91.04 |
4 |
6 |
2 |
74 |
264.329 |
2 |
↓
|
|