|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3E,5S,6S,8S,9R,10R,13S,14S,17S)-3-(2-aminoethoxyimino)-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,1
(3E,5S,6S,8S,9R,10R,13S,14S,17S)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
2.26 |
-43.42 |
5 |
5 |
1 |
90 |
365.538 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3E,5S,6S,8S,9S,10R,13S,14S,17S)-3-(2-aminoethoxyimino)-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,1
(3E,5S,6S,8S,9S,10R,13S,14S,17S)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
2.17 |
-43.45 |
5 |
5 |
1 |
90 |
365.538 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3E,5S,6S,8R,9R,10R,13S,14S,17S)-3-(2-aminoethoxyimino)-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,1
(3E,5S,6S,8R,9R,10R,13S,14S,17S)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
2.34 |
-43.1 |
5 |
5 |
1 |
90 |
365.538 |
3 |
↓
|
|
|
|
Analogs
-
12402552
-
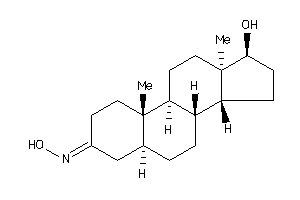
-
12402553
-
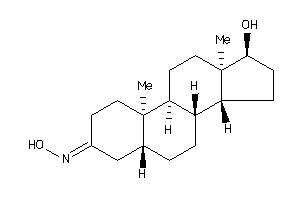
-
12402554
-
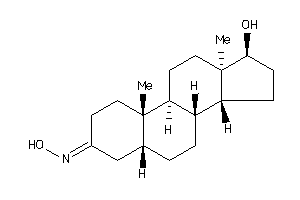
-
12877346
-
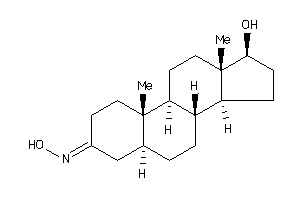
-
13146629
-
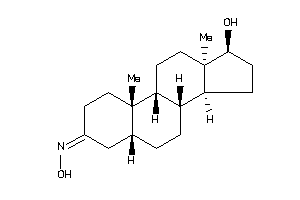
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5S,8R,9S,10R,13R,14R,17S)-17-hydroxy-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydr
(5S,8R,9S,10R,13R,14R,17S)-17-hy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
4.74 |
-6.24 |
2 |
3 |
0 |
53 |
305.462 |
0 |
↓
|
|
|
|
Analogs
-
12402553
-

-
12402554
-

-
12877346
-

-
13146629
-

-
13146631
-
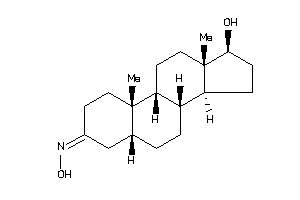
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5S,8R,9S,10S,13R,14R,17S)-17-hydroxy-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydr
(5S,8R,9S,10S,13R,14R,17S)-17-hy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
4.9 |
-5.3 |
2 |
3 |
0 |
53 |
305.462 |
0 |
↓
|
|
|
|
Analogs
-
12402554
-

-
12877346
-

-
13146629
-

-
13146631
-

-
4082983
-
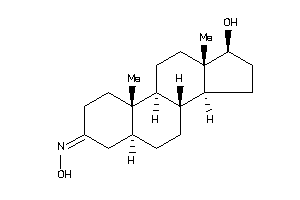
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5R,8R,9S,10R,13R,14R,17S)-17-hydroxy-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydr
(5R,8R,9S,10R,13R,14R,17S)-17-hy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
4.91 |
-6.27 |
2 |
3 |
0 |
53 |
305.462 |
0 |
↓
|
|
|
|
Analogs
-
12877346
-

-
13146629
-

-
13146631
-

-
4082983
-

-
12402551
-
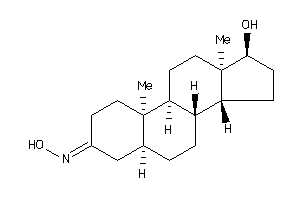
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5R,8R,9S,10S,13R,14R,17S)-17-hydroxy-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydr
(5R,8R,9S,10S,13R,14R,17S)-17-hy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
4.71 |
-5.25 |
2 |
3 |
0 |
53 |
305.462 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.09 |
9.57 |
-10.31 |
3 |
6 |
0 |
94 |
445.648 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.09 |
10.06 |
-10.71 |
3 |
6 |
0 |
94 |
445.648 |
6 |
↓
|
|
|
|
Analogs
-
38802343
-
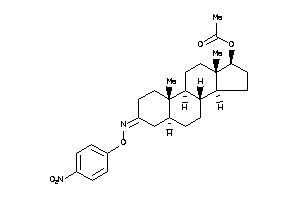
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.84 |
16.06 |
-12.5 |
0 |
7 |
0 |
94 |
503.039 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(Z)-[(5S,8R,9S,10S,13R,14R,17R)-14-hydroxy-10,13-dimethyl-17-(5-oxo-2H-furan-3-yl)-2,4,5,6,7,8,9,11
[(Z)-[(5S,8R,9S,10S,13R,14R,17R)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
8.57 |
-18.02 |
4 |
6 |
0 |
97 |
445.629 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(Z)-[(5R,8R,9S,10S,13R,14R,17R)-14-hydroxy-10,13-dimethyl-17-(5-oxo-2H-furan-3-yl)-2,4,5,6,7,8,9,11
[(Z)-[(5R,8R,9S,10S,13R,14R,17R)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
8.35 |
-18.25 |
4 |
6 |
0 |
97 |
445.629 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(Z)-[(5S,8S,9S,10S,13R,14R,17R)-14-hydroxy-10,13-dimethyl-17-(5-oxo-2H-furan-3-yl)-2,4,5,6,7,8,9,11
[(Z)-[(5S,8S,9S,10S,13R,14R,17R)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
8.37 |
-17.92 |
4 |
6 |
0 |
97 |
445.629 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(Z)-[(5R,8S,9S,10S,13R,14R,17R)-14-hydroxy-10,13-dimethyl-17-(5-oxo-2H-furan-3-yl)-2,4,5,6,7,8,9,11
[(Z)-[(5R,8S,9S,10S,13R,14R,17R)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
8.17 |
-18.24 |
4 |
6 |
0 |
97 |
445.629 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[(5S,8S,9R,10S,13R,14S,17S)-17-[(1R)-1,5-dimethylhexyl]-10,13-dimethyl-4,5,6,7,8,9,11,12,14,15,16,
1-[(5S,8S,9R,10S,13R,14S,17S)-17…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.81 |
17.7 |
-27.28 |
0 |
1 |
1 |
3 |
440.78 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[(5S,8S,9R,10S,13R,14S,17S)-17-[(1S)-1,5-dimethylhexyl]-10,13-dimethyl-4,5,6,7,8,9,11,12,14,15,16,
1-[(5S,8S,9R,10S,13R,14S,17S)-17…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.81 |
17.58 |
-27.73 |
0 |
1 |
1 |
3 |
440.78 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[(5S,8S,9R,10S,13R,14S,17R)-17-[(1R)-1,5-dimethylhexyl]-10,13-dimethyl-4,5,6,7,8,9,11,12,14,15,16,
1-[(5S,8S,9R,10S,13R,14S,17R)-17…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.81 |
17.83 |
-26.69 |
0 |
1 |
1 |
3 |
440.78 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[(5S,8S,9R,10S,13R,14S,17R)-17-[(1S)-1,5-dimethylhexyl]-10,13-dimethyl-4,5,6,7,8,9,11,12,14,15,16,
1-[(5S,8S,9R,10S,13R,14S,17R)-17…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.81 |
17.83 |
-26.86 |
0 |
1 |
1 |
3 |
440.78 |
5 |
↓
|
|