|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[(2E,7S,10R,13R)-13-[(4-hydroxyphenyl)methyl]-6,9,12-trioxo-7-propyl-17-oxa-5,8,11,14-tetrazabicyc
3-[(2E,7S,10R,13R)-13-[(4-hydrox…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4898 |
0.18 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
6.36 |
-76.68 |
6 |
11 |
0 |
173 |
566.655 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.97 |
5.25 |
-60.44 |
5 |
11 |
-1 |
169 |
565.647 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
650 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.23 |
1.11 |
-53.86 |
7 |
10 |
1 |
154 |
525.626 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.23 |
0.04 |
-13.38 |
6 |
10 |
0 |
149 |
524.618 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2E,7S,10R,13R)-13-benzyl-10-isopropyl-7-propyl-17-oxa-5,8,11,14-tetrazabicyclo[16.4.0]docosa-1(18),
(2E,7S,10R,13R)-13-benzyl-10-iso…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
78 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
8.78 |
-47.28 |
5 |
8 |
1 |
113 |
521.682 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.78 |
7.73 |
-8.83 |
4 |
8 |
0 |
109 |
520.674 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
2-[(2E,7S,10R,13R)-13-[(4-hydroxyphenyl)methyl]-10-isopropyl-6,9,12-trioxo-17-oxa-5,8,11,14-tetrazab
2-[(2E,7S,10R,13R)-13-[(4-hydrox…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2553 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.38 |
5.06 |
-78.99 |
6 |
11 |
0 |
173 |
552.628 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.38 |
3.92 |
-54.34 |
5 |
11 |
-1 |
169 |
551.62 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.38 |
3.07 |
-57.09 |
7 |
11 |
1 |
171 |
553.636 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
664 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.72 |
4.33 |
-106.84 |
9 |
10 |
2 |
161 |
567.731 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.72 |
3.2 |
-53.5 |
8 |
10 |
1 |
156 |
566.723 |
7 |
↓
|
|
|
|
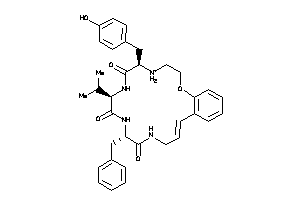
Analogs
-
36160717
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1389 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.21 |
4.28 |
-52.01 |
6 |
9 |
1 |
133 |
509.627 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.21 |
3.18 |
-11.46 |
5 |
9 |
0 |
129 |
508.619 |
3 |
↓
|
|
|
|
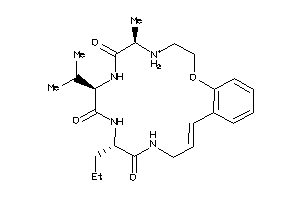
Analogs
-
40810757
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2E,7S,10R,13R)-10,13-diisopropyl-7-propyl-17-oxa-5,8,11,14-tetrazabicyclo[16.4.0]docosa-1(18),2,19,
(2E,7S,10R,13R)-10,13-diisopropy…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3566 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
6.8 |
-48.53 |
5 |
8 |
1 |
113 |
473.638 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.10 |
5.72 |
-8.24 |
4 |
8 |
0 |
109 |
472.63 |
4 |
↓
|
|
|
|
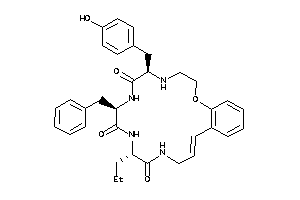
Analogs
-
36160713
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
196 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
4.68 |
-51.13 |
6 |
9 |
1 |
133 |
509.627 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.53 |
3.64 |
-11.97 |
5 |
9 |
0 |
129 |
508.619 |
4 |
↓
|
|
|
|
|
|
|
|
|
|
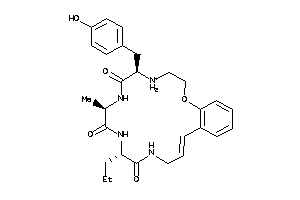
Analogs
-
36160687
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1524 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
7.74 |
-12.62 |
5 |
9 |
0 |
129 |
584.717 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.99 |
8.14 |
-50.8 |
6 |
9 |
1 |
133 |
585.725 |
6 |
↓
|
|
|
|
Analogs
-
36160672
-
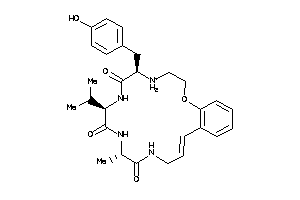
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
185 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.67 |
8.4 |
-52.93 |
6 |
9 |
1 |
133 |
585.725 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.67 |
7.23 |
-12.42 |
5 |
9 |
0 |
129 |
584.717 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.83 |
4.67 |
-118.71 |
8 |
9 |
2 |
141 |
503.688 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.83 |
3.63 |
-51.15 |
7 |
9 |
1 |
136 |
502.68 |
7 |
↓
|
|