|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2777 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.70 |
8.28 |
-55.42 |
5 |
9 |
1 |
121 |
553.724 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.70 |
5.22 |
-15.1 |
4 |
9 |
0 |
120 |
552.716 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3247 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.87 |
9.96 |
-47.04 |
5 |
8 |
1 |
113 |
529.746 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.87 |
8.72 |
-7.46 |
4 |
8 |
0 |
109 |
528.738 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
222 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.91 |
7.93 |
-63.12 |
5 |
9 |
1 |
125 |
567.751 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.91 |
6.85 |
-14.82 |
4 |
9 |
0 |
120 |
566.743 |
5 |
↓
|
|
|
|
Analogs
-
14210459
-
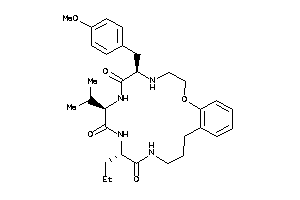
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
55 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.46 |
6.56 |
-49.06 |
6 |
9 |
1 |
133 |
539.697 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.46 |
5.32 |
-10.95 |
5 |
9 |
0 |
129 |
538.689 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1486 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.45 |
10.19 |
-51.84 |
5 |
8 |
1 |
113 |
537.725 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.45 |
8.92 |
-9.55 |
4 |
8 |
0 |
109 |
536.717 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
255 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.08 |
9.4 |
-48.06 |
6 |
9 |
1 |
129 |
562.735 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.08 |
8.09 |
-10.37 |
5 |
9 |
0 |
124 |
561.727 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
79 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.12 |
11.67 |
-47.95 |
5 |
8 |
1 |
113 |
573.758 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.12 |
10.42 |
-10.23 |
4 |
8 |
0 |
109 |
572.75 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.09 |
11.53 |
-47.22 |
5 |
8 |
1 |
113 |
573.758 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.09 |
10.22 |
-9.94 |
4 |
8 |
0 |
109 |
572.75 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6391 |
0.17 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
8.55 |
-15.42 |
4 |
10 |
0 |
137 |
580.726 |
5 |
↓
|
|