|
|
Analogs
-
36160961
-
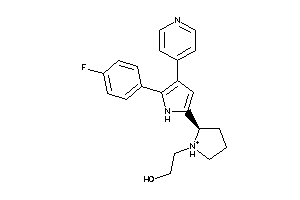
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
8.02 |
-40.73 |
3 |
4 |
1 |
53 |
352.433 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.12 |
8.5 |
-82.78 |
4 |
4 |
2 |
55 |
353.441 |
5 |
↓
|
|
|
|
Analogs
-
36160959
-
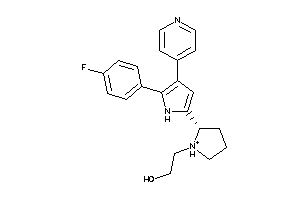
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
8.17 |
-40.55 |
3 |
4 |
1 |
53 |
352.433 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.12 |
8.84 |
-81.81 |
4 |
4 |
2 |
55 |
353.441 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
36160978
-
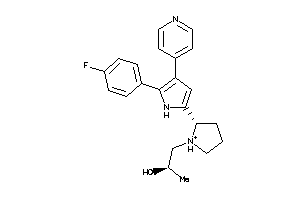
-
36160965
-
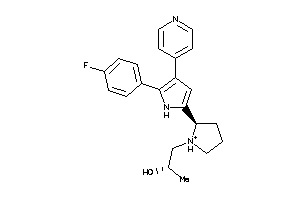
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
8.75 |
-40.44 |
3 |
4 |
1 |
53 |
366.46 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.48 |
9.23 |
-84.03 |
4 |
4 |
2 |
55 |
367.468 |
5 |
↓
|
|
|
|
Analogs
-
36160965
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
8.98 |
-40.53 |
3 |
4 |
1 |
53 |
366.46 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.48 |
9.46 |
-84.41 |
4 |
4 |
2 |
55 |
367.468 |
5 |
↓
|
|
|
|
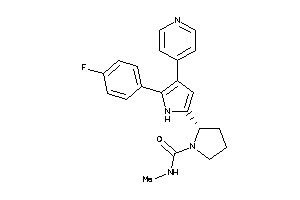
Analogs
-
36160990
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
8.56 |
-13.53 |
2 |
5 |
0 |
61 |
364.424 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.95 |
9.04 |
-39.47 |
3 |
5 |
1 |
62 |
365.432 |
3 |
↓
|
|
|
|
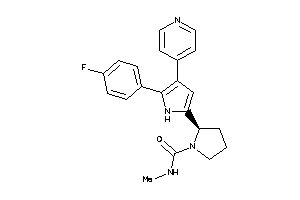
Analogs
-
36160983
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
8.55 |
-13.35 |
2 |
5 |
0 |
61 |
364.424 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.95 |
9.03 |
-40.5 |
3 |
5 |
1 |
62 |
365.432 |
3 |
↓
|
|
|
|
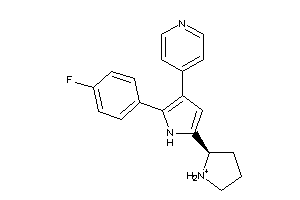
Analogs
-
36161016
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.51 |
8.55 |
-46.12 |
3 |
3 |
1 |
45 |
308.38 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.51 |
9.24 |
-87.87 |
4 |
3 |
2 |
47 |
309.388 |
3 |
↓
|
|
|
|
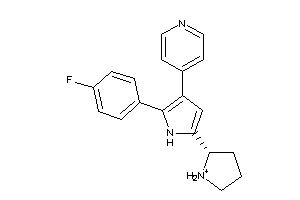
Analogs
-
36161012
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.51 |
8.61 |
-46.32 |
3 |
3 |
1 |
45 |
308.38 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.51 |
9.27 |
-87.81 |
4 |
3 |
2 |
47 |
309.388 |
3 |
↓
|
|
|
|
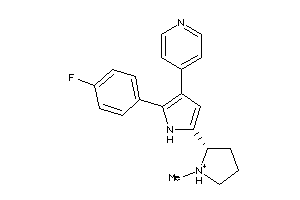
Analogs
-
36161045
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
10.6 |
-41.97 |
2 |
3 |
1 |
33 |
322.407 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.75 |
8.53 |
-5.85 |
1 |
3 |
0 |
32 |
321.399 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.75 |
11.08 |
-86.18 |
3 |
3 |
2 |
34 |
323.415 |
3 |
↓
|
|
|
|
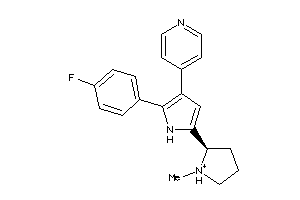
Analogs
-
36161040
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
10.6 |
-41.54 |
2 |
3 |
1 |
33 |
322.407 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.75 |
8.54 |
-5.86 |
1 |
3 |
0 |
32 |
321.399 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.75 |
11.07 |
-85.32 |
3 |
3 |
2 |
34 |
323.415 |
3 |
↓
|
|