|
|
Analogs
-
36161136
-
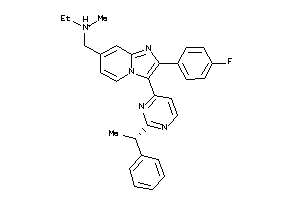
-
36161141
-
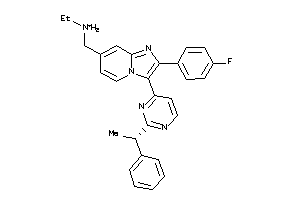
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.32 |
18 |
-51.51 |
1 |
5 |
1 |
48 |
480.611 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.32 |
18.71 |
-103.31 |
2 |
5 |
2 |
49 |
481.619 |
8 |
↓
|
|
|
|
Analogs
-
36161141
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.32 |
14.69 |
-57.9 |
2 |
5 |
1 |
60 |
438.53 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.32 |
13.24 |
-11.28 |
1 |
5 |
0 |
55 |
437.522 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.25 |
11.96 |
-13.3 |
1 |
6 |
0 |
67 |
453.521 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.25 |
12.56 |
-31.92 |
2 |
6 |
1 |
68 |
454.529 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.50 |
16.5 |
-53.63 |
2 |
5 |
1 |
60 |
480.611 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.52 |
14.5 |
-12.6 |
0 |
6 |
0 |
56 |
467.548 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.52 |
14.81 |
-34.95 |
1 |
6 |
1 |
57 |
468.556 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.68 |
13.79 |
-16.4 |
1 |
6 |
0 |
72 |
465.532 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.68 |
14.11 |
-40.64 |
2 |
6 |
1 |
73 |
466.54 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.94 |
12.81 |
-63.91 |
3 |
5 |
1 |
71 |
424.503 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.94 |
12.41 |
-11.6 |
2 |
5 |
0 |
69 |
423.495 |
5 |
↓
|
|
|
|
Analogs
-
36161131
-
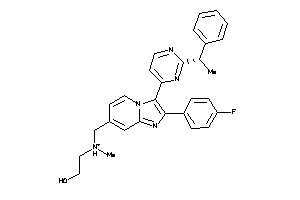
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
14.73 |
-52.72 |
2 |
6 |
1 |
68 |
496.61 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.31 |
15.47 |
-102.07 |
3 |
6 |
2 |
69 |
497.618 |
9 |
↓
|
|
|
|
Analogs
-
36161125
-
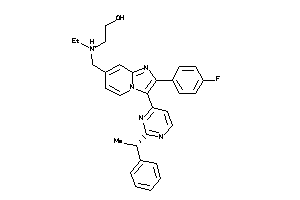
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
14.15 |
-52.06 |
2 |
6 |
1 |
68 |
482.583 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.93 |
11.73 |
-12.96 |
1 |
6 |
0 |
67 |
481.575 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.93 |
14.47 |
-104.39 |
3 |
6 |
2 |
69 |
483.591 |
8 |
↓
|
|
|
|
Analogs
-
36161141
-

-
36161080
-
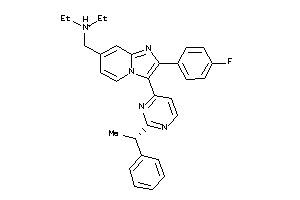
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.94 |
17.36 |
-52.84 |
1 |
5 |
1 |
48 |
466.584 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.94 |
18.09 |
-104.81 |
2 |
5 |
2 |
49 |
467.592 |
7 |
↓
|
|
|
|
Analogs
-
36161080
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q8MMZ8-1-E |
CGMP-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.69 |
15.53 |
-57.37 |
2 |
5 |
1 |
60 |
452.557 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.69 |
14.16 |
-12.23 |
1 |
5 |
0 |
55 |
451.549 |
7 |
↓
|
|