|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
990 |
0.23 |
Binding ≤ 10μM
|
|
SOAT2-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
990 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.03 |
16.57 |
-17.02 |
2 |
6 |
0 |
70 |
522.759 |
16 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.03 |
17 |
-29.55 |
3 |
6 |
1 |
71 |
523.767 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.83 |
19.19 |
-15.27 |
2 |
7 |
0 |
67 |
606.925 |
18 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.83 |
19.61 |
-32.1 |
3 |
7 |
1 |
69 |
607.933 |
18 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.29 |
Binding ≤ 10μM
|
|
SOAT2-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.79 |
17 |
-15.17 |
2 |
7 |
0 |
79 |
580.839 |
18 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.79 |
17.42 |
-36.64 |
3 |
7 |
1 |
81 |
581.847 |
18 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
340 |
0.22 |
Binding ≤ 10μM
|
|
SOAT2-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
340 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.14 |
15.17 |
-17.78 |
2 |
8 |
0 |
89 |
582.811 |
18 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.14 |
15.59 |
-33.96 |
3 |
8 |
1 |
90 |
583.819 |
18 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.21 |
Binding ≤ 10μM
|
|
SOAT2-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2400 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.08 |
18 |
-44.98 |
3 |
6 |
1 |
65 |
536.81 |
16 |
↓
|
|
Hi
High (pH 8-9.5)
|
8.08 |
15.69 |
-13.15 |
2 |
6 |
0 |
64 |
535.802 |
16 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.08 |
18.45 |
-99.76 |
4 |
6 |
2 |
67 |
537.818 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.83 |
18.34 |
-13.97 |
2 |
5 |
0 |
61 |
520.787 |
17 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.83 |
18.77 |
-32.17 |
3 |
5 |
1 |
62 |
521.795 |
17 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.74 |
18.4 |
-13.93 |
2 |
5 |
0 |
61 |
520.787 |
16 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.74 |
18.82 |
-31.92 |
3 |
5 |
1 |
62 |
521.795 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
910 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.50 |
15.26 |
-14.08 |
3 |
5 |
0 |
75 |
478.706 |
15 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.50 |
15.68 |
-32.49 |
4 |
5 |
1 |
76 |
479.714 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.35 |
16.74 |
-14.39 |
2 |
5 |
0 |
61 |
492.733 |
15 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.35 |
17.16 |
-32.71 |
3 |
5 |
1 |
62 |
493.741 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
34 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.78 |
19.48 |
-12.06 |
1 |
5 |
0 |
58 |
521.771 |
18 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.78 |
19.9 |
-31.44 |
2 |
5 |
1 |
59 |
522.779 |
18 |
↓
|
|
|
|
Analogs
-
6363752
-
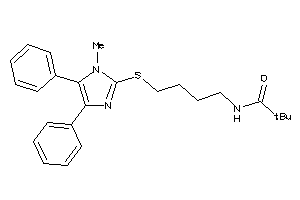
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.51 |
19 |
-13.15 |
1 |
4 |
0 |
49 |
505.772 |
16 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.51 |
19.45 |
-36.11 |
2 |
4 |
1 |
50 |
506.78 |
16 |
↓
|
|
|
|
|
|
|
Analogs
-
44651826
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
6.14 |
-49.38 |
4 |
3 |
1 |
56 |
250.325 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.56 |
5.74 |
-8.95 |
3 |
3 |
0 |
55 |
249.317 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.24 |
6.92 |
-52.01 |
4 |
3 |
1 |
56 |
264.352 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.24 |
6.61 |
-7.22 |
3 |
3 |
0 |
55 |
263.344 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.24 |
7.31 |
-116.22 |
5 |
3 |
2 |
58 |
265.36 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.24 |
6.64 |
-46.11 |
4 |
3 |
1 |
56 |
264.352 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.24 |
6.34 |
-7.4 |
3 |
3 |
0 |
55 |
263.344 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.24 |
7.31 |
-115.7 |
5 |
3 |
2 |
58 |
265.36 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
7.75 |
-50.96 |
4 |
3 |
1 |
56 |
278.379 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.75 |
7.45 |
-7.11 |
3 |
3 |
0 |
55 |
277.371 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.75 |
8.12 |
-115.96 |
5 |
3 |
2 |
58 |
279.387 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
7.46 |
-44.98 |
4 |
3 |
1 |
56 |
278.379 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.75 |
7.14 |
-7.14 |
3 |
3 |
0 |
55 |
277.371 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.75 |
8.12 |
-115.49 |
5 |
3 |
2 |
58 |
279.387 |
4 |
↓
|
|
|
|
Analogs
-
13281668
-
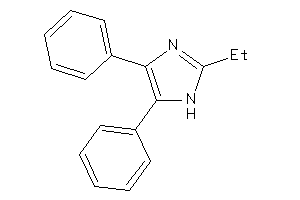
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
7.64 |
-48.14 |
4 |
3 |
1 |
56 |
278.379 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.63 |
8.1 |
-100.12 |
5 |
3 |
2 |
58 |
279.387 |
5 |
↓
|
|
|
|
Analogs
-
36876337
-
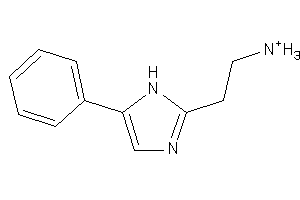
-
13281668
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.36 |
6.87 |
-49.82 |
4 |
3 |
1 |
56 |
264.352 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.36 |
7.31 |
-110.15 |
5 |
3 |
2 |
58 |
265.36 |
4 |
↓
|
|
|
|
Analogs
-
36877334
-

-
44651826
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
8.02 |
-43.6 |
3 |
3 |
1 |
45 |
264.352 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.94 |
6.58 |
-7.06 |
2 |
3 |
0 |
41 |
263.344 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.68 |
19.13 |
-12.94 |
1 |
4 |
0 |
49 |
505.772 |
17 |
↓
|
|
Mid
Mid (pH 6-8)
|
8.68 |
19.54 |
-37.74 |
2 |
4 |
1 |
50 |
506.78 |
17 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.17 |
7.81 |
-13.83 |
1 |
3 |
0 |
46 |
248.285 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
5.62 |
-8.75 |
2 |
3 |
0 |
49 |
250.301 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.71 |
6.11 |
-28.42 |
3 |
3 |
1 |
50 |
251.309 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.58 |
9.23 |
-9.28 |
1 |
4 |
0 |
47 |
358.388 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.58 |
9.82 |
-41.47 |
2 |
4 |
1 |
49 |
359.396 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.58 |
9.24 |
-9.26 |
1 |
4 |
0 |
47 |
358.388 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.58 |
9.82 |
-38.09 |
2 |
4 |
1 |
49 |
359.396 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.23 |
11.68 |
-56.56 |
0 |
5 |
-1 |
67 |
335.383 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.23 |
12.28 |
-46.67 |
1 |
5 |
0 |
68 |
336.391 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.19 |
9.39 |
-49.08 |
3 |
4 |
1 |
55 |
322.432 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.19 |
9.97 |
-93.71 |
4 |
4 |
2 |
56 |
323.44 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
6.32 |
-9.18 |
2 |
3 |
0 |
49 |
250.301 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.56 |
6.76 |
-38.59 |
3 |
3 |
1 |
50 |
251.309 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
7.02 |
-16.3 |
1 |
6 |
0 |
73 |
389.452 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.65 |
7.55 |
-44.71 |
2 |
6 |
1 |
74 |
390.46 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
10.84 |
-43.4 |
2 |
5 |
1 |
52 |
380.512 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.94 |
8.66 |
-9.8 |
1 |
5 |
0 |
51 |
379.504 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.94 |
11.39 |
-80.34 |
3 |
5 |
2 |
53 |
381.52 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
10.85 |
-44.63 |
2 |
5 |
1 |
52 |
380.512 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.94 |
8.69 |
-9.75 |
1 |
5 |
0 |
51 |
379.504 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.94 |
11.39 |
-80.57 |
3 |
5 |
2 |
53 |
381.52 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
7.62 |
-13.6 |
1 |
6 |
0 |
73 |
385.489 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.77 |
8.23 |
-43.97 |
2 |
6 |
1 |
74 |
386.497 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
8.49 |
-14.03 |
1 |
3 |
0 |
38 |
268.291 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.40 |
9 |
-38.62 |
2 |
3 |
1 |
39 |
269.299 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.17 |
12.08 |
-47.11 |
2 |
3 |
1 |
34 |
344.841 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.17 |
12.61 |
-97.14 |
3 |
3 |
2 |
36 |
345.849 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.90 |
11.82 |
-11.9 |
0 |
4 |
0 |
51 |
331.419 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.90 |
12.48 |
-33.93 |
1 |
4 |
1 |
52 |
332.427 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.90 |
12.07 |
-12.1 |
0 |
4 |
0 |
51 |
331.419 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.90 |
12.74 |
-33.56 |
1 |
4 |
1 |
52 |
332.427 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
9.29 |
-57.69 |
1 |
4 |
-1 |
69 |
263.276 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.80 |
9.68 |
-48.26 |
2 |
4 |
0 |
70 |
264.284 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
11.43 |
-93.57 |
3 |
4 |
2 |
44 |
351.494 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.03 |
11.01 |
-46.97 |
2 |
4 |
1 |
42 |
350.486 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
8.55 |
-17.85 |
0 |
5 |
0 |
61 |
374.437 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.89 |
9.09 |
-45.65 |
1 |
5 |
1 |
62 |
375.445 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.98 |
14.06 |
-44.08 |
1 |
3 |
1 |
22 |
338.45 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.98 |
11.92 |
-7.54 |
0 |
3 |
0 |
21 |
337.442 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.98 |
14.62 |
-80.69 |
2 |
3 |
2 |
24 |
339.458 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.98 |
14.06 |
-44.41 |
1 |
3 |
1 |
22 |
338.45 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.98 |
11.81 |
-7.45 |
0 |
3 |
0 |
21 |
337.442 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.98 |
14.63 |
-81.28 |
2 |
3 |
2 |
24 |
339.458 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
8.37 |
-19 |
0 |
5 |
0 |
61 |
374.437 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.87 |
8.88 |
-44.85 |
1 |
5 |
1 |
62 |
375.445 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.85 |
10.39 |
-12.89 |
1 |
4 |
0 |
47 |
351.425 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.85 |
11.56 |
-31.83 |
2 |
4 |
1 |
48 |
352.433 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.85 |
10.76 |
-13.08 |
1 |
4 |
0 |
47 |
351.425 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.85 |
11.5 |
-31.66 |
2 |
4 |
1 |
48 |
352.433 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
9.62 |
-13.64 |
1 |
4 |
0 |
47 |
336.435 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.59 |
10.17 |
-36.72 |
2 |
4 |
1 |
49 |
337.443 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
9.68 |
-12.39 |
1 |
4 |
0 |
47 |
336.435 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.59 |
10.23 |
-36.18 |
2 |
4 |
1 |
49 |
337.443 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
11.66 |
-9.88 |
0 |
3 |
0 |
27 |
302.377 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.93 |
12.17 |
-35.16 |
1 |
3 |
1 |
28 |
303.385 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.00 |
11.45 |
-16.8 |
1 |
5 |
0 |
56 |
377.488 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.00 |
11.99 |
-39.89 |
2 |
5 |
1 |
57 |
378.496 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
9.21 |
-9.97 |
2 |
3 |
0 |
49 |
302.377 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.65 |
9.73 |
-40.02 |
3 |
3 |
1 |
50 |
303.385 |
2 |
↓
|
|