|
|
Analogs
-
4942480
-
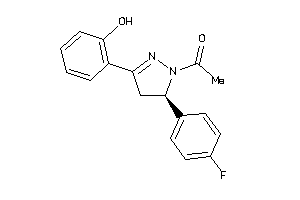
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABP1-2-E |
Diamine Oxidase (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.32 |
Binding ≤ 10μM
|
|
AOC2-1-E |
Retina-specific Amine Oxidase, Copper Containing (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
|
AOC3-2-E |
Amine Oxidase, Copper Containing (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
7.41 |
-10.8 |
1 |
4 |
0 |
53 |
294.354 |
2 |
↓
|
|
|
|
Analogs
-
4942480
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABP1-2-E |
Diamine Oxidase (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.32 |
Binding ≤ 10μM
|
|
AOC2-1-E |
Retina-specific Amine Oxidase, Copper Containing (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
|
AOC3-2-E |
Amine Oxidase, Copper Containing (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
7.43 |
-10.56 |
1 |
4 |
0 |
53 |
294.354 |
2 |
↓
|
|
|
|
Analogs
-
14951055
-
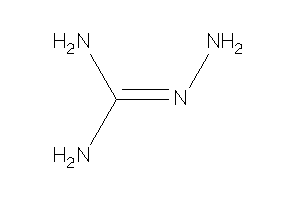
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABP1-3-E |
Diamine Oxidase (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
140 |
1.92 |
Binding ≤ 10μM |
|
NOS1-4-E |
Nitric-oxide Synthase, Brain (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
5900 |
1.46 |
Binding ≤ 10μM
|
|
NOS2-6-E |
Nitric Oxide Synthase, Inducible (cluster #6 Of 9), Eukaryotic |
Eukaryotes |
3900 |
1.51 |
Binding ≤ 10μM
|
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
1.46 |
Functional ≤ 10μM
|
|
NOS2-3-E |
Nitric Oxide Synthase, Inducible (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4500 |
1.50 |
Functional ≤ 10μM
|
|
Z80418-8-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #8 Of 9), Other |
Other |
50 |
2.04 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.27 |
-7.4 |
-28.95 |
7 |
4 |
1 |
89 |
75.095 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABP1-1-E |
Diamine Oxidase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
160 |
0.95 |
Binding ≤ 10μM
|
|
AMO-1-E |
Amine Oxidase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
710 |
0.86 |
Binding ≤ 10μM
|
|
AOC3-1-E |
Amine Oxidase, Copper Containing (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.71 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.16 |
-0.24 |
-48.33 |
4 |
3 |
1 |
53 |
138.194 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.16 |
0.05 |
-97.36 |
5 |
3 |
2 |
54 |
139.202 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABP1-1-E |
Diamine Oxidase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
300 |
1.01 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.54 |
-1.19 |
-48.29 |
5 |
3 |
1 |
67 |
124.167 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.54 |
-0.91 |
-97.81 |
6 |
3 |
2 |
68 |
125.175 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABP1-1-E |
Diamine Oxidase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5000 |
0.62 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.20 |
1.41 |
-48.18 |
4 |
3 |
1 |
53 |
164.232 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.20 |
1.7 |
-99.94 |
5 |
3 |
2 |
54 |
165.24 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABP1-1-E |
Diamine Oxidase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.80 |
Binding ≤ 10μM
|
|
AMO-1-E |
Amine Oxidase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9300 |
0.64 |
Binding ≤ 10μM
|
|
AOC3-1-E |
Amine Oxidase, Copper Containing (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.73 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.21 |
0.57 |
-47.9 |
4 |
3 |
1 |
53 |
152.221 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.21 |
0.85 |
-97.77 |
5 |
3 |
2 |
54 |
153.229 |
3 |
↓
|
|