|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 49 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A-3-E |
Alpha-1a Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.62 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2600 |
0.65 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2600 |
0.65 |
Binding ≤ 10μM
|
|
ADA2A-3-E |
Alpha-2a Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
750 |
0.71 |
Binding ≤ 10μM
|
|
ADA2B-4-E |
Alpha-2b Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
78 |
0.83 |
Binding ≤ 10μM
|
|
ADA2C-3-E |
Alpha-2c Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
750 |
0.71 |
Binding ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2700 |
0.65 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.61 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.70 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.80 |
Functional ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
150 |
0.80 |
Functional ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
52 |
0.85 |
Functional ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
52 |
0.85 |
Functional ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
52 |
0.85 |
Functional ≤ 10μM
|
|
DRD2-1-E |
Dopamine D2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
7300 |
0.60 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 1), Other |
Other |
200 |
0.78 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
1500 |
0.68 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.04 |
-4.68 |
-50.09 |
6 |
4 |
1 |
88 |
170.188 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.04 |
-5.06 |
-7.63 |
5 |
4 |
0 |
87 |
169.18 |
2 |
↓
|
|
|
|
Analogs
-
12650844
-
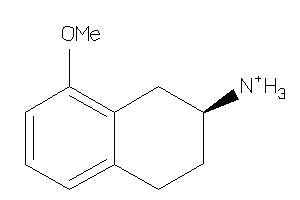
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
53 |
0.78 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
902 |
0.65 |
Binding ≤ 10μM |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.62 |
Functional ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1700 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.50 |
3.57 |
-44.79 |
3 |
2 |
1 |
37 |
178.255 |
1 |
↓
|
|
|
|
Analogs
-
2567336
-
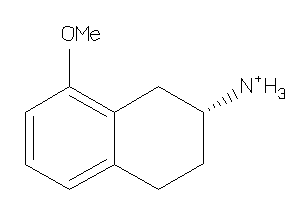
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
53 |
0.78 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
902 |
0.65 |
Binding ≤ 10μM |
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.62 |
Functional ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1700 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.50 |
3.6 |
-44.82 |
3 |
2 |
1 |
37 |
178.255 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 39 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
683 |
0.66 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.61 |
Functional ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2200 |
0.61 |
Functional ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
334 |
0.70 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.50 |
3.59 |
-47.71 |
3 |
2 |
1 |
37 |
178.255 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
683 |
0.66 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.61 |
Functional ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2200 |
0.61 |
Functional ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
334 |
0.70 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.50 |
3.59 |
-47.78 |
3 |
2 |
1 |
37 |
178.255 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|