|
|
Analogs
-
8071079
-
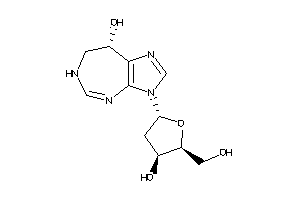
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
250 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.08 |
-3.94 |
-19.13 |
4 |
8 |
0 |
112 |
268.273 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.08 |
-3.46 |
-31.4 |
5 |
8 |
1 |
113 |
269.281 |
2 |
↓
|
|
|
|
Analogs
-
44608135
-
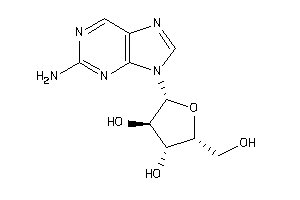
-
44608136
-
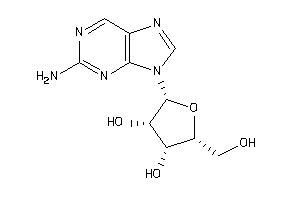
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.40 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.46 |
-3.79 |
-15.03 |
5 |
9 |
0 |
140 |
267.245 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.39 |
Binding ≤ 10μM
|
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.63 |
Binding ≤ 10μM |
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.74 |
-4.14 |
-17.12 |
3 |
9 |
0 |
133 |
265.229 |
6 |
↓
|
|
|
|
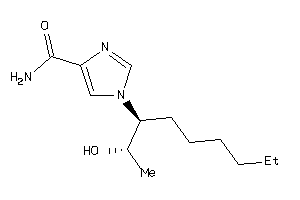
Analogs
-
12405523
-
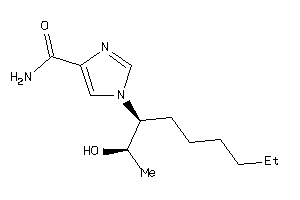
-
34633627
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.72 |
-2.47 |
-11.17 |
3 |
5 |
0 |
81 |
253.346 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.82 |
-2.15 |
-38.48 |
4 |
5 |
1 |
82 |
254.354 |
8 |
↓
|
|
|
|
Analogs
-
12405523
-

-
34633627
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.72 |
3.58 |
-9.31 |
3 |
5 |
0 |
81 |
253.346 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.72 |
4.11 |
-40.91 |
4 |
5 |
1 |
82 |
254.354 |
8 |
↓
|
|
|
|
|
|
|
|
|
|
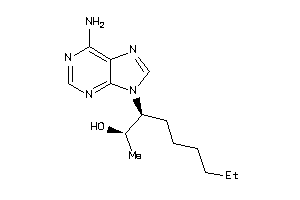
Analogs
-
1614354
-
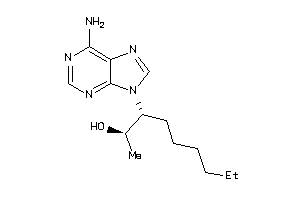
-
1614355
-
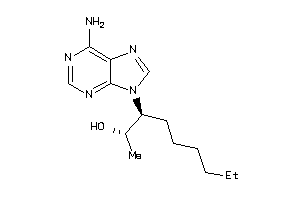
-
1614356
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Binding ≤ 10μM
|
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Binding ≤ 10μM
|
|
PDE2A-1-E |
Phosphodiesterase 2A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
635 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
-3.11 |
-8.94 |
3 |
6 |
0 |
89 |
277.372 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.37 |
-2.92 |
-34.6 |
4 |
6 |
1 |
91 |
278.38 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.97 |
-2.92 |
-34.6 |
4 |
6 |
1 |
91 |
278.38 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.38 |
Binding ≤ 10μM
|
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.63 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.74 |
-4.14 |
-17.04 |
3 |
9 |
0 |
133 |
265.229 |
6 |
↓
|
|
|
|
Analogs
-
1558334
-
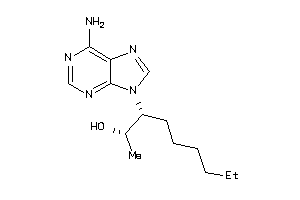
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
455 |
0.44 |
Binding ≤ 10μM
|
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.57 |
Binding ≤ 10μM
|
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
80 |
0.50 |
Binding ≤ 10μM
|
|
PDE2A-1-E |
Phosphodiesterase 2A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
6.86 |
-10.9 |
3 |
6 |
0 |
90 |
277.372 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.98 |
7.13 |
-33.95 |
4 |
6 |
1 |
91 |
278.38 |
7 |
↓
|
|
|
|
Analogs
-
16951808
-
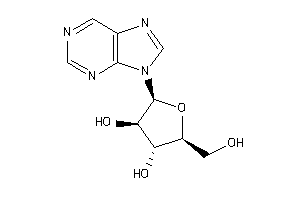
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.40 |
Binding ≤ 10μM |
|
CDD-1-E |
Cytidine Deaminase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.39 |
Binding ≤ 10μM
|
|
Z80583-7-O |
Vero (Kidney Cells) (cluster #7 Of 7), Other |
Other |
1 |
0.70 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.27 |
-3.14 |
-14.27 |
3 |
8 |
0 |
114 |
252.23 |
2 |
↓
|
|