|
|
Analogs
-
4942480
-
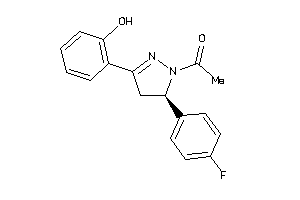
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABP1-2-E |
Diamine Oxidase (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.32 |
Binding ≤ 10μM
|
|
AOC2-1-E |
Retina-specific Amine Oxidase, Copper Containing (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
|
AOC3-2-E |
Amine Oxidase, Copper Containing (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
7.41 |
-10.8 |
1 |
4 |
0 |
53 |
294.354 |
2 |
↓
|
|
|
|
Analogs
-
4942480
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABP1-2-E |
Diamine Oxidase (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.32 |
Binding ≤ 10μM
|
|
AOC2-1-E |
Retina-specific Amine Oxidase, Copper Containing (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
|
AOC3-2-E |
Amine Oxidase, Copper Containing (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
7.43 |
-10.56 |
1 |
4 |
0 |
53 |
294.354 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
400 |
0.53 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
5000 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
6.13 |
-7.8 |
1 |
3 |
0 |
34 |
232.258 |
3 |
↓
|
|
|
|
Analogs
-
32225469
-
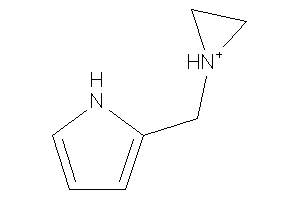
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-3-E |
Monoamine Oxidase A (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
5 |
1.06 |
Binding ≤ 10μM
|
|
AOFB-3-E |
Monoamine Oxidase B (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
20 |
0.98 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.06 |
3.29 |
-5.06 |
1 |
2 |
0 |
19 |
148.209 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-1-E |
Monoamine Oxidase A (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
300 |
0.91 |
Binding ≤ 10μM
|
|
AOFB-1-E |
Monoamine Oxidase B (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
620 |
0.87 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.81 |
2.15 |
-4.45 |
2 |
2 |
0 |
28 |
134.182 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-1-E |
Monoamine Oxidase A (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
20 |
0.72 |
Binding ≤ 10μM
|
|
AOFB-1-E |
Monoamine Oxidase B (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
700 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
6.83 |
-43.61 |
3 |
2 |
1 |
32 |
201.293 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-1-E |
Monoamine Oxidase A (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
350 |
0.65 |
Binding ≤ 10μM
|
|
AOFB-1-E |
Monoamine Oxidase B (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
7200 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
-0.92 |
-42.36 |
3 |
2 |
1 |
32 |
187.266 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
31 |
0.48 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.50 |
-1.23 |
-10.87 |
2 |
3 |
0 |
41 |
329.856 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
31 |
0.48 |
Binding ≤ 10μM
|
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
50 |
0.46 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.54 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.50 |
-1.23 |
-10.9 |
2 |
3 |
0 |
41 |
329.856 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-3-E |
Monoamine Oxidase A (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
27 |
0.59 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
50 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
7.37 |
-10.97 |
2 |
3 |
0 |
42 |
293.442 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-3-E |
Monoamine Oxidase A (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
27 |
0.59 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
50 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
7.37 |
-10.99 |
2 |
3 |
0 |
42 |
293.442 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
50 |
0.51 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
10 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
8.99 |
-10.65 |
2 |
3 |
0 |
42 |
301.44 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
50 |
0.51 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
10 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
8.99 |
-10.66 |
2 |
3 |
0 |
42 |
301.44 |
3 |
↓
|
|
|
|
Analogs
-
5187207
-
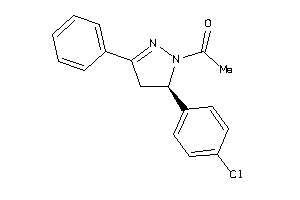
-
1077164
-
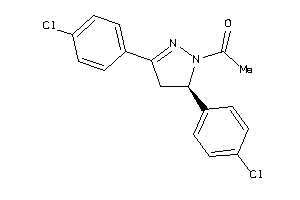
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6000 |
0.35 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6000 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.60 |
1.74 |
-8.4 |
0 |
3 |
0 |
32 |
298.773 |
2 |
↓
|
|
|
|
Analogs
-
5187204
-
-
1077164
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6000 |
0.35 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6000 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.60 |
1.72 |
-8.41 |
0 |
3 |
0 |
32 |
298.773 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
590 |
0.51 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8000 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.87 |
6.08 |
-9.17 |
1 |
3 |
0 |
34 |
232.258 |
3 |
↓
|
|
|
|
Analogs
-
25103609
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
360 |
0.50 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
450 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.16 |
7.91 |
-10.1 |
0 |
3 |
0 |
25 |
246.285 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6 |
0.52 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
2400 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.22 |
9.82 |
-12.09 |
2 |
3 |
0 |
42 |
333.819 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
41 |
0.52 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
8.83 |
-10.35 |
2 |
3 |
0 |
42 |
321.858 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
41 |
0.52 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
8.83 |
-10.37 |
2 |
3 |
0 |
42 |
321.858 |
3 |
↓
|
|
|
|
Analogs
-
3887002
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-3-E |
Monoamine Oxidase A (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
50 |
0.57 |
Binding ≤ 10μM
|
|
AOFB-3-E |
Monoamine Oxidase B (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
20 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.54 |
-2.03 |
-12.14 |
2 |
4 |
0 |
54 |
277.374 |
3 |
↓
|
|
|
|
Analogs
-
3887001
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-3-E |
Monoamine Oxidase A (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
50 |
0.57 |
Binding ≤ 10μM
|
|
AOFB-3-E |
Monoamine Oxidase B (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
20 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.54 |
-2.03 |
-12.16 |
2 |
4 |
0 |
54 |
277.374 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
50 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
9.99 |
-11.95 |
2 |
3 |
0 |
42 |
313.401 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
50 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
9.98 |
-11.95 |
2 |
3 |
0 |
42 |
313.401 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 94 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-1-E |
Monoamine Oxidase B (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
720 |
0.78 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.21 |
4.28 |
-43.63 |
3 |
1 |
1 |
28 |
148.229 |
0 |
↓
|
|
|
|
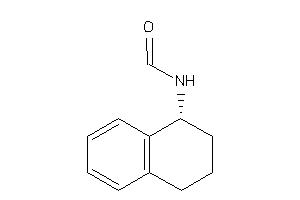
Analogs
-
4582529
-
-
5689234
-
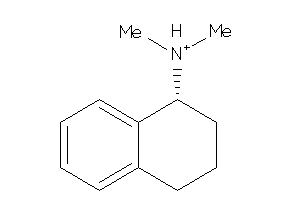
-
4204787
-
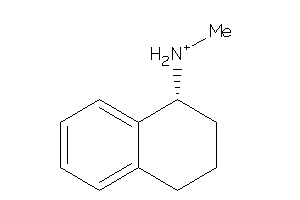
Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-1-E |
Monoamine Oxidase B (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1400 |
0.68 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.56 |
6.08 |
-36.9 |
2 |
1 |
1 |
17 |
162.256 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6 |
0.52 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.22 |
9.8 |
-12.19 |
2 |
3 |
0 |
42 |
333.819 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6 |
0.55 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
960 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.30 |
9.92 |
-10.78 |
0 |
3 |
0 |
25 |
278.355 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.35 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
10000 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.50 |
1.8 |
-9.64 |
0 |
4 |
0 |
41 |
342.826 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.35 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
10000 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.50 |
1.81 |
-9.63 |
0 |
4 |
0 |
41 |
342.826 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
10000 |
0.41 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
320 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
7.03 |
-4.72 |
2 |
2 |
0 |
28 |
222.291 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
130 |
0.54 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
230 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
6.44 |
-9.5 |
2 |
3 |
0 |
45 |
236.274 |
2 |
↓
|
|
|
|
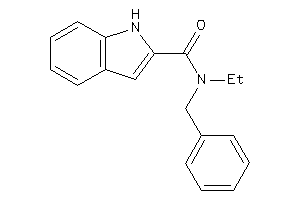
Analogs
-
35127898
-
-
35395843
-
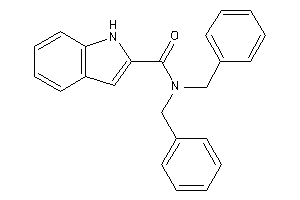
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
70 |
0.50 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1600 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
8.79 |
-12.88 |
1 |
3 |
0 |
36 |
264.328 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4 |
0.56 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.64 |
9.69 |
-10.79 |
1 |
3 |
0 |
36 |
278.355 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
5 |
0.55 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
800 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.91 |
8.12 |
-9.51 |
2 |
3 |
0 |
45 |
278.355 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
170 |
0.47 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
190 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
7.34 |
-10.22 |
2 |
3 |
0 |
45 |
264.328 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.46 |
5.96 |
-35.86 |
3 |
3 |
1 |
50 |
265.336 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4 |
0.62 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
94 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
0.68 |
-10.14 |
1 |
3 |
0 |
36 |
250.301 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
15 |
0.55 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
21 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.60 |
9.08 |
-9.95 |
0 |
3 |
0 |
25 |
264.328 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
30 |
0.55 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
120 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
6.56 |
-8.21 |
2 |
3 |
0 |
45 |
250.301 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.06 |
5.18 |
-34.59 |
3 |
3 |
1 |
50 |
251.309 |
3 |
↓
|
|
|
|
Analogs
-
7909010
-
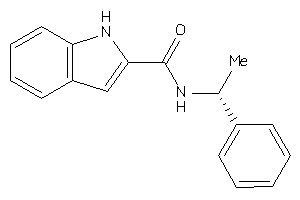
-
37842166
-
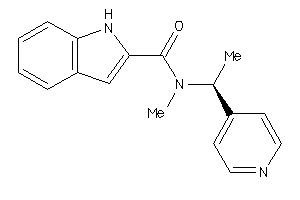
-
37842167
-
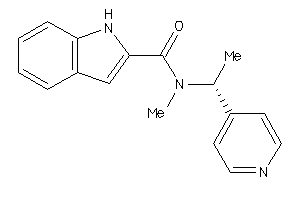
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
250 |
0.46 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
500 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
7.4 |
-8.75 |
2 |
3 |
0 |
45 |
264.328 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.62 |
6.08 |
-33.29 |
3 |
3 |
1 |
50 |
265.336 |
3 |
↓
|
|
|
|
Analogs
-
37842166
-

-
37842167
-

-
7909008
-
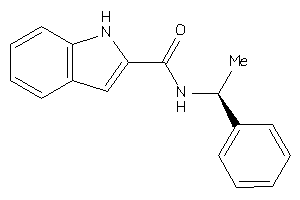
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
250 |
0.46 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
500 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
7.4 |
-8.75 |
2 |
3 |
0 |
45 |
264.328 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.62 |
6.07 |
-33.26 |
3 |
3 |
1 |
50 |
265.336 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
7244 |
0.33 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1148 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
2.67 |
-8.62 |
0 |
3 |
0 |
32 |
296.345 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
7244 |
0.33 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1148 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
2.67 |
-8.6 |
0 |
3 |
0 |
32 |
296.345 |
3 |
↓
|
|
|
|
Analogs
-
5063541
-
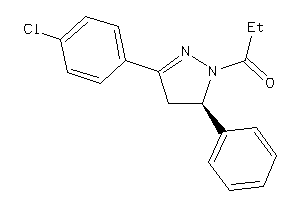
-
366785
-
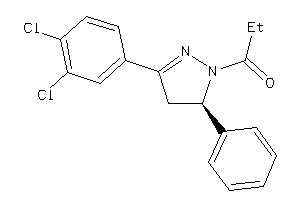
-
366787
-
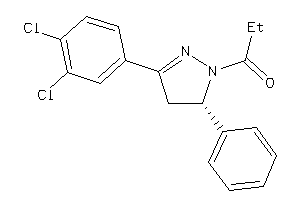
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
501 |
0.40 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1288 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.44 |
1.78 |
-8.53 |
0 |
3 |
0 |
32 |
312.8 |
3 |
↓
|
|
|
|
Analogs
-
366785
-

-
366787
-

-
612523
-
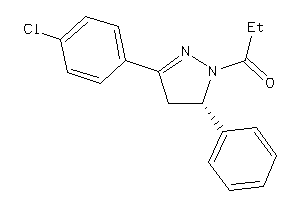
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-2-E |
Monoamine Oxidase A (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
501 |
0.40 |
Binding ≤ 10μM
|
|
AOFB-2-E |
Monoamine Oxidase B (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1288 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.44 |
1.79 |
-8.54 |
0 |
3 |
0 |
32 |
312.8 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFB-6-E |
Monoamine Oxidase B (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
5000 |
0.62 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.35 |
3.17 |
-43.08 |
3 |
2 |
1 |
37 |
186.662 |
4 |
↓
|
|
|
|
Analogs
-
22056030
-
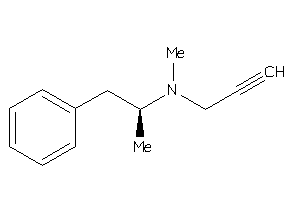
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AOFA-1-E |
Monoamine Oxidase A (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3800 |
0.54 |
Binding ≤ 10μM
|
|
AOFB-1-E |
Monoamine Oxidase B (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
970 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.64 |
6.48 |
-3.01 |
0 |
1 |
0 |
3 |
187.286 |
4 |
↓
|
|