|
|
|
|
|
Analogs
-
6096530
-
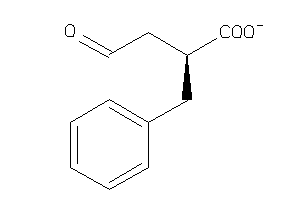
-
6096592
-
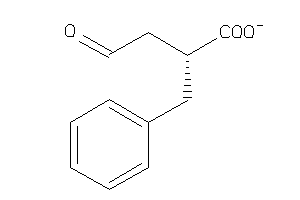
Draw
Identity
99%
90%
80%
70%
Vendors
And 30 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CBPA1-1-E |
Carboxypeptidase A1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.60 |
Binding ≤ 10μM
|
|
CBPA1-1-E |
Carboxypeptidase A1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
450 |
0.59 |
Binding ≤ 10μM
|
|
CBPA3-1-E |
Mast Cell Carboxypeptidase A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
450 |
0.59 |
Binding ≤ 10μM
|
|
CBPB1-2-E |
Carboxypeptidase B (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.47 |
Binding ≤ 10μM
|
|
CBPB2-1-E |
Carboxypeptidase B2 Isoform A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.25 |
6.05 |
-127.16 |
0 |
4 |
-2 |
80 |
206.197 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CBPB1-2-E |
Carboxypeptidase B (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4500 |
0.31 |
Binding ≤ 10μM
|
|
Q3T905-1-E |
Carboxypeptidase B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
1.22 |
-11.52 |
0 |
3 |
0 |
33 |
334.444 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CBPB1-3-E |
Carboxypeptidase B (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.29 |
Binding ≤ 10μM
|
|
Q3T905-1-E |
Carboxypeptidase B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.69 |
6.57 |
-13.19 |
1 |
6 |
0 |
73 |
444.969 |
8 |
↓
|
|
|
|
Analogs
-
4521543
-
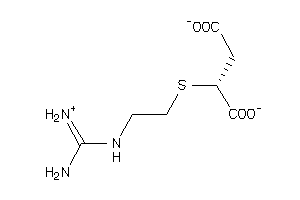
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CBPB1-1-E |
Carboxypeptidase B (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.55 |
-4.81 |
-100.74 |
5 |
7 |
-1 |
143 |
234.257 |
8 |
↓
|
|
|
|
Analogs
-
2384679
-
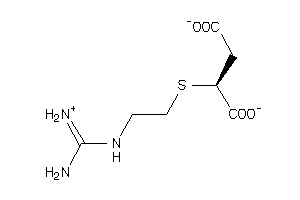
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CBPB1-1-E |
Carboxypeptidase B (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.55 |
-4.81 |
-100.73 |
5 |
7 |
-1 |
143 |
234.257 |
8 |
↓
|
|
|
|
Analogs
-
1223733
-
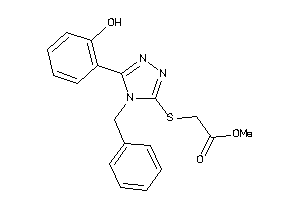
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CBPB1-3-E |
Carboxypeptidase B (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7400 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.70 |
3.22 |
-14.66 |
0 |
6 |
0 |
66 |
397.5 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CBPB1-3-E |
Carboxypeptidase B (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
9100 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.91 |
8.97 |
-14.29 |
1 |
6 |
0 |
81 |
397.481 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CBPA1-1-E |
Carboxypeptidase A1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.35 |
Binding ≤ 10μM
|
|
CBPB1-3-E |
Carboxypeptidase B (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7500 |
0.31 |
Binding ≤ 10μM
|
|
Q3T905-1-E |
Carboxypeptidase B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.09 |
3.36 |
-11.34 |
1 |
5 |
0 |
76 |
346.795 |
5 |
↓
|
|
|
|
|