|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
14 |
0.30 |
Binding ≤ 10μM
|
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM |
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
8 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.50 |
16.04 |
-72.01 |
2 |
6 |
1 |
64 |
514.685 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.29 |
Binding ≤ 10μM |
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4500 |
0.19 |
Binding ≤ 10μM |
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.30 |
Functional ≤ 10μM |
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.45 |
12.09 |
-51.07 |
1 |
7 |
1 |
63 |
560.685 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.45 |
9.97 |
-8.57 |
0 |
7 |
0 |
62 |
559.677 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.45 |
12.06 |
-45.59 |
1 |
7 |
1 |
63 |
560.685 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1415 |
0.24 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.25 |
Binding ≤ 10μM
|
|
CCR1-1-E |
C-C Chemokine Receptor Type 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.21 |
Binding ≤ 10μM
|
|
CCR2-1-E |
C-C Chemokine Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.21 |
Binding ≤ 10μM
|
|
CCR3-1-E |
C-C Chemokine Receptor Type 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.33 |
Binding ≤ 10μM
|
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.25 |
Binding ≤ 10μM
|
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.21 |
Binding ≤ 10μM
|
|
CXCR2-1-E |
Interleukin-8 Receptor B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.21 |
Binding ≤ 10μM
|
|
Q9WTR4-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
611 |
0.26 |
Binding ≤ 10μM
|
|
SC6A2-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
611 |
0.26 |
Binding ≤ 10μM
|
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
718 |
0.25 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2350 |
0.23 |
Binding ≤ 10μM
|
|
CCR3-1-E |
C-C Chemokine Receptor Type 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.33 |
Functional ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
240 |
0.27 |
ADME/T ≤ 10μM
|
|
DRD2-17-E |
Dopamine D2 Receptor (cluster #17 Of 24), Eukaryotic |
Eukaryotes |
321 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.40 |
14.84 |
-55.42 |
3 |
6 |
1 |
76 |
487.665 |
7 |
↓
|
|
|
|
Analogs
-
9686365
-
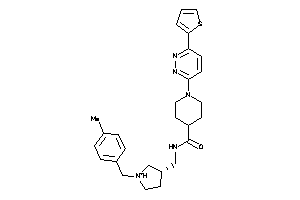
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5800 |
0.22 |
Binding ≤ 10μM
|
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.56 |
-2.76 |
-53.06 |
2 |
6 |
1 |
62 |
476.67 |
7 |
↓
|
|
|
|
Analogs
-
9686362
-
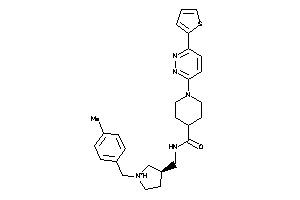
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5800 |
0.22 |
Binding ≤ 10μM
|
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.56 |
-2.68 |
-48.89 |
2 |
6 |
1 |
62 |
476.67 |
7 |
↓
|
|
|
|
Analogs
-
13720371
-
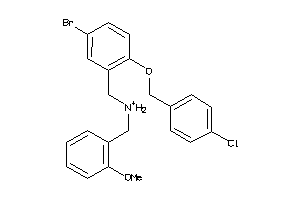
-
16480099
-
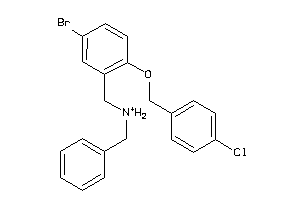
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCR5-2-E |
C-C Chemokine Receptor Type 5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1100 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.94 |
-0.6 |
-37.98 |
2 |
2 |
1 |
25 |
355.683 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCR5-2-E |
C-C Chemokine Receptor Type 5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1700 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.19 |
7.02 |
-45.41 |
3 |
2 |
1 |
37 |
327.629 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCR5-2-E |
C-C Chemokine Receptor Type 5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
8.57 |
-31.46 |
5 |
5 |
1 |
85 |
348.224 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
2.55 |
-42.69 |
1 |
4 |
1 |
38 |
495.687 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Binding ≤ 10μM |
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
1 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
11.23 |
-50.6 |
1 |
7 |
1 |
63 |
534.647 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.12 |
9.14 |
-9.26 |
0 |
7 |
0 |
62 |
533.639 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.93 |
11.38 |
-50.51 |
3 |
7 |
1 |
74 |
352.466 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCR2-1-E |
C-C Chemokine Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.29 |
Functional ≤ 10μM
|
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
10.64 |
-52.24 |
3 |
10 |
1 |
106 |
468.582 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.73 |
8.1 |
-27.52 |
2 |
10 |
0 |
105 |
467.574 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.82 |
15.76 |
-54.49 |
2 |
6 |
1 |
64 |
464.659 |
9 |
↓
|
|
|
|
Analogs
-
33727462
-
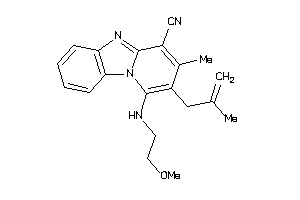
-
33727463
-
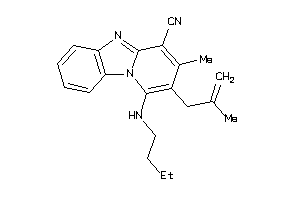
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.22 |
2.99 |
-54.46 |
2 |
5 |
1 |
57 |
390.555 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCR1-1-E |
C-C Chemokine Receptor Type 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
615 |
0.29 |
Binding ≤ 10μM
|
|
Q9JLY8-1-E |
C-C Chemokine Receptor Type 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
59 |
0.34 |
Binding ≤ 10μM
|
|
CCR1-1-E |
C-C Chemokine Receptor Type 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.27 |
Functional ≤ 10μM
|
|
CCR4-1-E |
C-C Chemokine Receptor Type 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.35 |
Functional ≤ 10μM
|
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
5.76 |
-18.99 |
3 |
7 |
0 |
88 |
434.899 |
6 |
↓
|
|