|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPC-1-B |
Beta-lactamase (cluster #1 Of 6), Bacterial |
Bacteria |
6 |
0.46 |
Binding ≤ 10μM
|
|
CP51-1-B |
Sterol 14-alpha Demethylase (cluster #1 Of 2), Bacterial |
Bacteria |
200 |
0.38 |
Binding ≤ 10μM
|
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
82 |
0.40 |
Binding ≤ 10μM
|
|
CP19A-3-E |
Cytochrome P450 19A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
CP51A-1-E |
Cytochrome P450 51 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
130 |
0.39 |
Binding ≤ 10μM
|
|
KCNA3-1-E |
Voltage-gated Potassium Channel Subunit Kv1.3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.29 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
3020 |
0.31 |
Binding ≤ 10μM
|
|
KCNN4-1-E |
Intermediate Conductance Calcium-activated Potassium Channel Protein 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.40 |
Binding ≤ 10μM
|
|
MDHM-1-E |
Malate Dehydrogenase, Mitochondrial (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6700 |
0.29 |
Functional ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4800 |
0.30 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.41 |
ADME/T ≤ 10μM
|
|
Q96W81-1-F |
14-alpha Sterol Demethylase (cluster #1 Of 1), Fungal |
Fungi |
103 |
0.39 |
Binding ≤ 10μM
|
|
Q4WNT5-1-O |
14-alpha Sterol Demethylase Cyp51A (cluster #1 Of 1), Other |
Other |
4790 |
0.30 |
Binding ≤ 10μM
|
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
6540 |
0.29 |
Functional ≤ 10μM
|
|
Z50425-9-O |
Plasmodium Falciparum (cluster #9 Of 22), Other |
Other |
60 |
0.40 |
Functional ≤ 10μM
|
|
Z50426-1-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #1 Of 9), Other |
Other |
250 |
0.37 |
Functional ≤ 10μM
|
|
Z80682-8-O |
A549 (Lung Carcinoma Cells) (cluster #8 Of 11), Other |
Other |
5100 |
0.30 |
Functional ≤ 10μM
|
|
Z80951-2-O |
NIH3T3 (Fibroblasts) (cluster #2 Of 4), Other |
Other |
630 |
0.35 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
3000 |
0.31 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.47 |
13.43 |
-5.78 |
0 |
2 |
0 |
18 |
344.845 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.47 |
13.99 |
-28.18 |
1 |
2 |
1 |
19 |
345.853 |
4 |
↓
|
|
|
|
Analogs
-
13102942
-
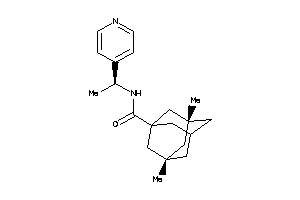
-
13102943
-
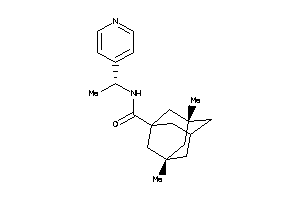
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
820 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.35 |
7.62 |
-8.13 |
1 |
3 |
0 |
42 |
284.403 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.35 |
7.9 |
-42.09 |
2 |
3 |
1 |
43 |
285.411 |
3 |
↓
|
|
|
|
Analogs
-
13102942
-

-
13102943
-

-
44836482
-
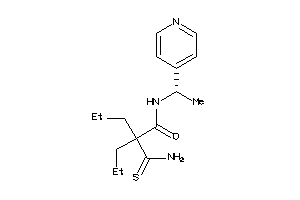
-
44836486
-
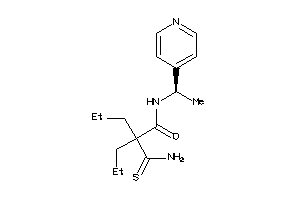
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
430 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.35 |
7.62 |
-8.14 |
1 |
3 |
0 |
42 |
284.403 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.35 |
7.9 |
-42.07 |
2 |
3 |
1 |
43 |
285.411 |
3 |
↓
|
|
|
|
Analogs
-
13119547
-
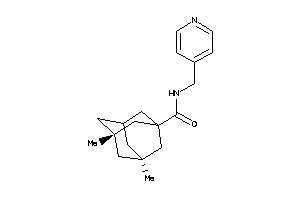
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7700 |
0.36 |
Binding ≤ 10μM
|
|
CP19A-2-E |
Cytochrome P450 19A1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1500 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
6.79 |
-7.41 |
1 |
3 |
0 |
42 |
270.376 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.80 |
7.24 |
-41.4 |
2 |
3 |
1 |
43 |
271.384 |
3 |
↓
|
|
|
|
Analogs
-
897385
-
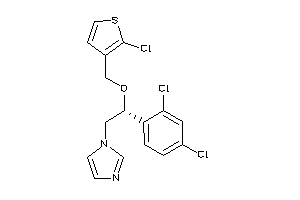
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
505 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.83 |
12.68 |
-34.29 |
1 |
3 |
1 |
28 |
388.727 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.83 |
12.17 |
-6.65 |
0 |
3 |
0 |
27 |
387.719 |
6 |
↓
|
|
|
|
Analogs
-
608101
-
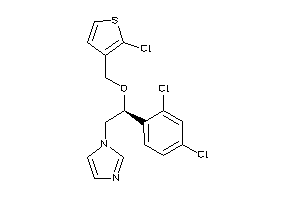
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
505 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.83 |
12.13 |
-33.31 |
1 |
3 |
1 |
28 |
388.727 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.83 |
11.61 |
-6.8 |
0 |
3 |
0 |
27 |
387.719 |
6 |
↓
|
|
|
|
Analogs
-
5356791
-
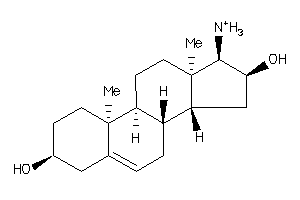
-
5356829
-
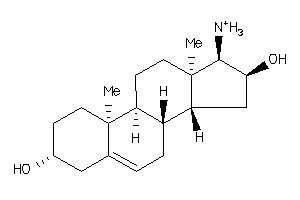
-
5359847
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
17-(1-aminoethyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthr
17-(1-aminoethyl)-10,13-dimethyl…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9800 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
-2.73 |
-45.41 |
4 |
2 |
1 |
47 |
318.525 |
1 |
↓
|
|
|
|
Analogs
-
5356791
-

-
5356829
-

-
5359847
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
17-(1-aminoethyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthr
17-(1-aminoethyl)-10,13-dimethyl…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9800 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
-2.03 |
-42.99 |
4 |
2 |
1 |
47 |
318.525 |
1 |
↓
|
|
|
|
Analogs
-
5356791
-

-
5356829
-

-
5359847
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
17-(1-aminoethyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthr
17-(1-aminoethyl)-10,13-dimethyl…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9800 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
-2.78 |
-43.82 |
4 |
2 |
1 |
47 |
318.525 |
1 |
↓
|
|
|
|
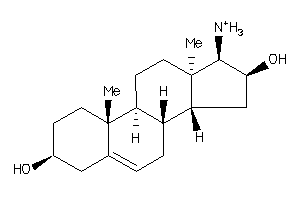
Analogs
-
4451747
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
17-(1-aminoethyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthr
17-(1-aminoethyl)-10,13-dimethyl…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9800 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
-2.35 |
-43.03 |
4 |
2 |
1 |
47 |
318.525 |
1 |
↓
|
|
|
|
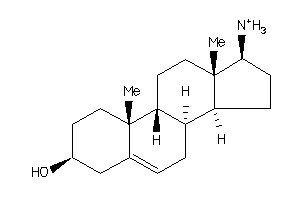
Analogs
-
4532104
-
-
4532105
-
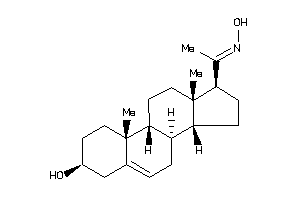
-
13759938
-
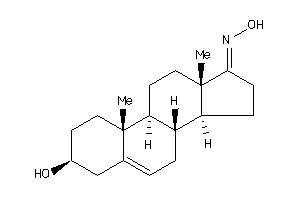
-
33938215
-
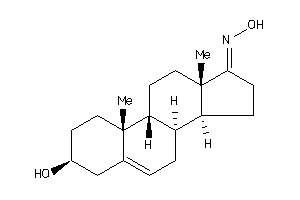
-
33938217
-
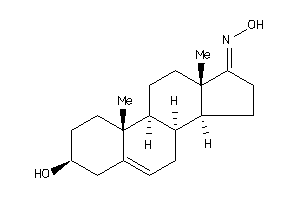
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-(3-hydroxy-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-17
1-(3-hydroxy-10,13-dimethyl-2,3,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
570 |
0.36 |
Binding ≤ 10μM
|
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1650 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.44 |
6.65 |
-4.75 |
2 |
3 |
0 |
53 |
331.5 |
1 |
↓
|
|
|
|
Analogs
-
13759938
-

-
33938215
-

-
33938217
-

-
4532102
-
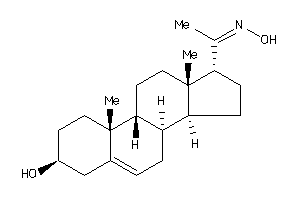
-
4532103
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-(3-hydroxy-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-17
1-(3-hydroxy-10,13-dimethyl-2,3,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
570 |
0.36 |
Binding ≤ 10μM
|
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1650 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.44 |
6.61 |
-4.44 |
2 |
3 |
0 |
53 |
331.5 |
1 |
↓
|
|
|
|
Analogs
-
3814414
-
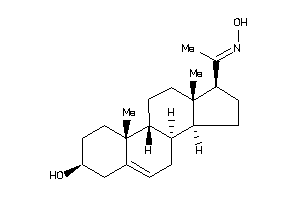
Draw
Identity
99%
90%
80%
70%
Popular Name:
17-(1-hydroxyethyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenant
17-(1-hydroxyethyl)-10,13-dimeth…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
720 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
-1.83 |
-2.97 |
2 |
2 |
0 |
40 |
318.501 |
1 |
↓
|
|
|
|
Analogs
-
3814414
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
17-(1-hydroxyethyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenant
17-(1-hydroxyethyl)-10,13-dimeth…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
720 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
-1.51 |
-2.75 |
2 |
2 |
0 |
40 |
318.501 |
1 |
↓
|
|
|
|
Analogs
-
3814414
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
17-(1-hydroxyethyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenant
17-(1-hydroxyethyl)-10,13-dimeth…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
190 |
0.41 |
Binding ≤ 10μM
|
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
720 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
-1.67 |
-2.64 |
2 |
2 |
0 |
40 |
318.501 |
1 |
↓
|
|
|
|
Analogs
-
3814414
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
17-(1-hydroxyethyl)-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenant
17-(1-hydroxyethyl)-10,13-dimeth…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
190 |
0.41 |
Binding ≤ 10μM
|
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
720 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
-2.1 |
-2.77 |
2 |
2 |
0 |
40 |
318.501 |
1 |
↓
|
|
|
|
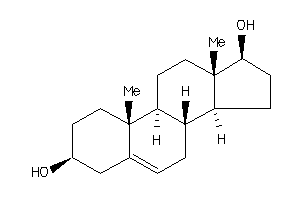
Analogs
-
103270
-
-
490793
-
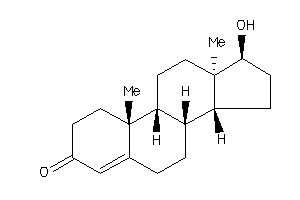
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8R,9R,10R,13S,14S,17S)-17-(1-hydroxyethyl)-10,13-dimethyl-1,2,6,7,8,9,11,12,14,15,16,17-dodecahydro
(8R,9R,10R,13S,14S,17S)-17-(1-hy…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
490 |
0.38 |
Binding ≤ 10μM
|
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2840 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
8.26 |
-6.42 |
1 |
2 |
0 |
37 |
316.485 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
968234
-
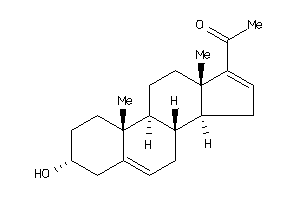
-
968235
-
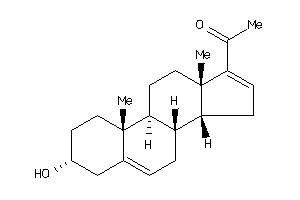
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-(3-hydroxy-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15-decahydro-1H-cyclopenta[a]phenanthren-17-yl)etha
1-(3-hydroxy-10,13-dimethyl-2,3,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
510 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
0.47 |
-8.51 |
1 |
2 |
0 |
37 |
314.469 |
1 |
↓
|
|
|
|
Analogs
-
968234
-

-
968235
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
1-(3-hydroxy-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15-decahydro-1H-cyclopenta[a]phenanthren-17-yl)etha
1-(3-hydroxy-10,13-dimethyl-2,3,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
510 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
1.01 |
-8.14 |
1 |
2 |
0 |
37 |
314.469 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C11B1-2-E |
Cytochrome P450 11B1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1268 |
0.49 |
Binding ≤ 10μM
|
|
C11B2-1-E |
Cytochrome P450 11B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.67 |
Binding ≤ 10μM
|
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1085 |
0.49 |
Binding ≤ 10μM
|
|
CP19A-1-E |
Cytochrome P450 19A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6045 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
9.2 |
-4.81 |
0 |
1 |
0 |
13 |
221.303 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.15 |
9.48 |
-34.74 |
1 |
1 |
1 |
14 |
222.311 |
1 |
↓
|
|
|
|
Analogs
-
26392960
-
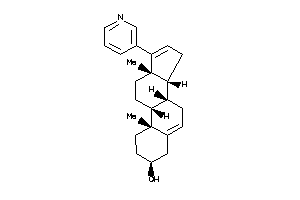
-
44830820
-
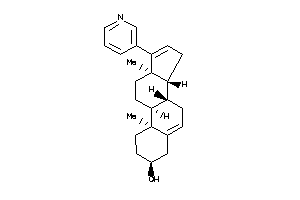
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.42 |
-0.59 |
-5.05 |
1 |
2 |
0 |
33 |
349.518 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
790 |
0.36 |
Binding ≤ 10μM
|
|
CP51A-1-E |
Cytochrome P450 51 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
342 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.97 |
15.01 |
-36.26 |
1 |
2 |
1 |
19 |
311.408 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.97 |
14.05 |
-7.54 |
0 |
2 |
0 |
18 |
310.4 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 32 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
790 |
0.36 |
Binding ≤ 10μM
|
|
CP51A-1-E |
Cytochrome P450 51 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
342 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.97 |
14.99 |
-36.29 |
1 |
2 |
1 |
19 |
311.408 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.97 |
14.04 |
-7.59 |
0 |
2 |
0 |
18 |
310.4 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
5359129
-
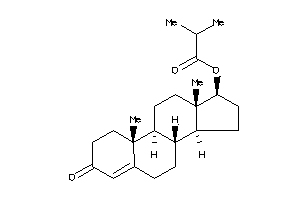
-
3875080
-
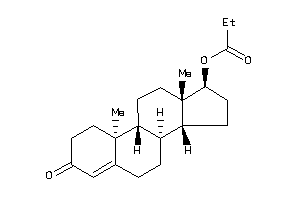
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
11.9 |
-10.74 |
0 |
3 |
0 |
43 |
344.495 |
3 |
↓
|
|
|
|
Analogs
-
3875080
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
11.95 |
-9.99 |
0 |
3 |
0 |
43 |
344.495 |
3 |
↓
|
|
|
|
Analogs
-
3875080
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
56 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
11.72 |
-10.5 |
0 |
3 |
0 |
43 |
344.495 |
3 |
↓
|
|
|
|
Analogs
-
643138
-
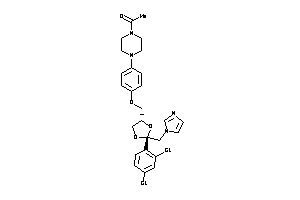
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.77 |
13.08 |
-47.53 |
1 |
8 |
1 |
70 |
532.448 |
7 |
↓
|
|
|
|
|
|
|
Analogs
-
10272298
-
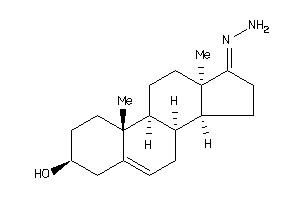
-
10272299
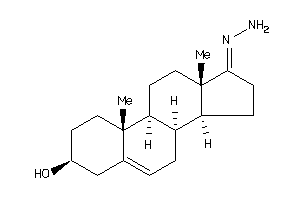
-
-
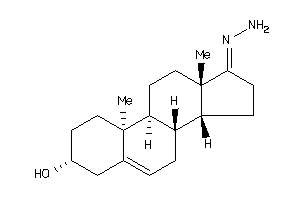
12719842
-
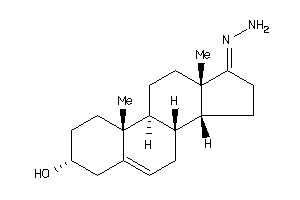
-
12719843
-
-
12719845
-
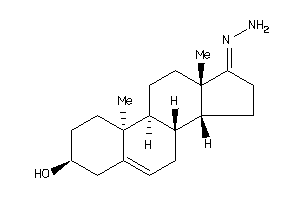
Draw
Identity
99%
90%
80%
70%
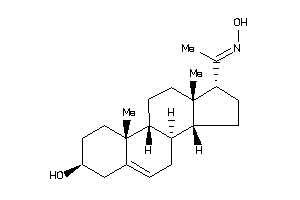
Popular Name:
17-hydrazono-10,13-dimethyl-1,2,3,4,7,8,9,11,12,14,15,16-dodecahydrocyclopenta[a]phenanthren-3-ol
17-hydrazono-10,13-dimethyl-1,2,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6480 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
5.63 |
-4.4 |
3 |
3 |
0 |
59 |
302.462 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.40 |
5.53 |
-38.31 |
4 |
3 |
1 |
60 |
303.47 |
0 |
↓
|
|
|
|
Analogs
-
12719842
-

-
12719843
-

-
12719845
-

-
12719847
-
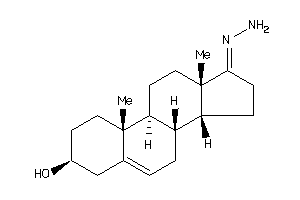
-
10272296
-
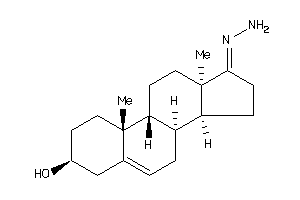
Draw
Identity
99%
90%
80%
70%
Popular Name:
17-hydrazono-10,13-dimethyl-1,2,3,4,7,8,9,11,12,14,15,16-dodecahydrocyclopenta[a]phenanthren-3-ol
17-hydrazono-10,13-dimethyl-1,2,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6480 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
5.6 |
-4.55 |
3 |
3 |
0 |
59 |
302.462 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.40 |
5.48 |
-38.83 |
4 |
3 |
1 |
60 |
303.47 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,8R,9S,10R,13S,14S)-3-hydroxy-10,13-dimethyl-1,2,3,4,7,8,9,11,12,14,15,16-dodecahydrocyclopenta[a
(3S,8R,9S,10R,13S,14S)-3-hydroxy…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6480 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
5.5 |
-4.49 |
3 |
3 |
0 |
59 |
302.462 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,8R,9R,10R,13S,14S)-3-hydroxy-10,13-dimethyl-1,2,3,4,7,8,9,11,12,14,15,16-dodecahydrocyclopenta[a
(3S,8R,9R,10R,13S,14S)-3-hydroxy…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6480 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
5.59 |
-4.53 |
3 |
3 |
0 |
59 |
302.462 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.40 |
5.4 |
-41.22 |
4 |
3 |
1 |
60 |
303.47 |
0 |
↓
|
|
|
|
Analogs
-
4532103
-

-
4532104
-

-
4532105
-

-
13759938
-

-
33938215
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
1-(3-hydroxy-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-17
1-(3-hydroxy-10,13-dimethyl-2,3,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
570 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.44 |
6.6 |
-4.97 |
2 |
3 |
0 |
53 |
331.5 |
1 |
↓
|
|
|
|
Analogs
-
4532105
-

-
13759938
-

-
33938215
-

-
33938217
-

-
4532102
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
1-(3-hydroxy-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-17
1-(3-hydroxy-10,13-dimethyl-2,3,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
570 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.44 |
5.99 |
-5.35 |
2 |
3 |
0 |
53 |
331.5 |
1 |
↓
|
|
|
|
Analogs
-
753542
-
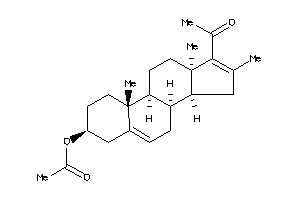
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1890 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.51 |
4.03 |
-10.32 |
0 |
3 |
0 |
43 |
356.506 |
3 |
↓
|
|
|
|
Analogs
-
753542
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1890 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.51 |
4.09 |
-9.65 |
0 |
3 |
0 |
43 |
356.506 |
3 |
↓
|
|
|
|
Analogs
-
753542
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1890 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.51 |
3.9 |
-10.61 |
0 |
3 |
0 |
43 |
356.506 |
3 |
↓
|
|
|
|
Analogs
-
753542
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(17-acetyl-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15-decahydro-1H-cyclopenta[a]phenanthren-3-yl)
(17-acetyl-10,13-dimethyl-2,3,4,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1890 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.51 |
3.89 |
-9.95 |
0 |
3 |
0 |
43 |
356.506 |
3 |
↓
|
|
|
|
Analogs
-
3982458
-
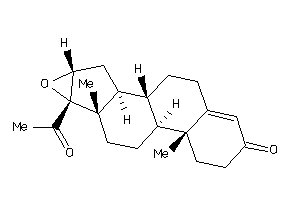
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
0.98 |
-9.45 |
0 |
3 |
0 |
46 |
328.452 |
1 |
↓
|
|
|
|
Analogs
-
4543816
-
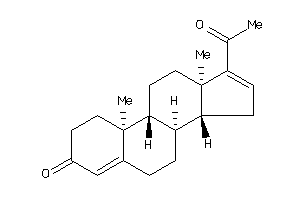
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9R,10R,13S,14S)-17-acetyl-10,13-dimethyl-1,2,6,7,8,9,11,12,14,15-decahydrocyclopenta[a]phenanthr
(8S,9R,10R,13S,14S)-17-acetyl-10…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1770 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.62 |
10.54 |
-10.85 |
0 |
2 |
0 |
34 |
312.453 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
-2.03 |
-8.27 |
2 |
5 |
0 |
62 |
429.973 |
8 |
↓
|
|
|
|
Analogs
-
643143
-
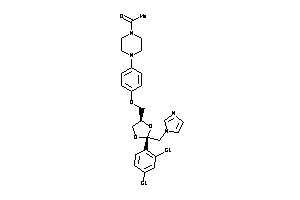
-
643153
-
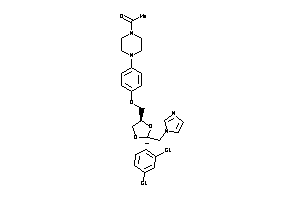
-
3872994
-
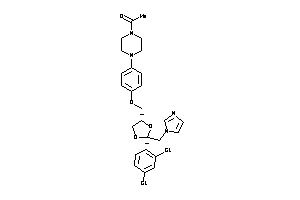
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C11B1-1-E |
Cytochrome P450 11B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
224 |
0.26 |
Binding ≤ 10μM
|
|
C11B2-2-E |
Cytochrome P450 11B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
81 |
0.28 |
Binding ≤ 10μM
|
|
CP11A-2-E |
Cytochrome P450 11A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5400 |
0.20 |
Binding ≤ 10μM
|
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Binding ≤ 10μM
|
|
CP19A-1-E |
Cytochrome P450 19A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
900 |
0.24 |
Binding ≤ 10μM
|
|
CP21A-1-E |
Cytochrome P450 21 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9010 |
0.20 |
Binding ≤ 10μM
|
|
CP24A-1-E |
Cytochrome P450 24A1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
520 |
0.24 |
Binding ≤ 10μM
|
|
CP3A4-1-E |
Cytochrome P450 3A4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
57 |
0.28 |
Binding ≤ 10μM
|
|
CP51A-1-E |
Cytochrome P450 51 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
65 |
0.28 |
Binding ≤ 10μM
|
|
CP7A1-1-E |
Cytochrome P450 7A1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2400 |
0.22 |
Binding ≤ 10μM
|
|
GNRHR-1-E |
Gonadotropin-releasing Hormone Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
811 |
0.24 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5720 |
0.20 |
Binding ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.23 |
Binding ≤ 10μM
|
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2780 |
0.22 |
Functional ≤ 10μM
|
|
MDR1-1-E |
P-glycoprotein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4800 |
0.21 |
Functional ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3800 |
0.21 |
Functional ≤ 10μM
|
|
CP24A-1-E |
Cytochrome P450 24A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
520 |
0.24 |
ADME/T ≤ 10μM |
|
CP2A2-1-E |
Cytochrome P450 2A2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
543 |
0.24 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
786 |
0.24 |
ADME/T ≤ 10μM
|
|
Z102121-2-O |
Trichophyton Mentagrophytes (cluster #2 Of 3), Other |
Other |
200 |
0.26 |
Functional ≤ 10μM
|
|
Z50274-1-O |
Aspergillus Flavus (cluster #1 Of 2), Other |
Other |
2800 |
0.22 |
Functional ≤ 10μM
|
|
Z50275-1-O |
Aspergillus Niger (cluster #1 Of 2), Other |
Other |
6000 |
0.20 |
Functional ≤ 10μM
|
|
Z50408-1-O |
Issatchenkia Orientalis (cluster #1 Of 2), Other |
Other |
400 |
0.25 |
Functional ≤ 10μM
|
|
Z50416-1-O |
Aspergillus Fumigatus (cluster #1 Of 3), Other |
Other |
3200 |
0.21 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.20 |
Functional ≤ 10μM
|
|
Z50436-1-O |
Filobasidiella Neoformans (cluster #1 Of 3), Other |
Other |
20 |
0.30 |
Functional ≤ 10μM
|
|
Z50442-1-O |
Candida Albicans (cluster #1 Of 4), Other |
Other |
300 |
0.25 |
Functional ≤ 10μM
|
|
Z50443-1-O |
Candida Glabrata (cluster #1 Of 1), Other |
Other |
700 |
0.24 |
Functional ≤ 10μM
|
|
Z50444-1-O |
Candida Parapsilosis (cluster #1 Of 2), Other |
Other |
40 |
0.29 |
Functional ≤ 10μM
|
|
Z50446-1-O |
Candida Tropicalis (cluster #1 Of 2), Other |
Other |
40 |
0.29 |
Functional ≤ 10μM
|
|
Z50452-1-O |
Trichophyton Rubrum (cluster #1 Of 2), Other |
Other |
20 |
0.30 |
Functional ≤ 10μM
|
|
Z50459-1-O |
Leishmania Donovani (cluster #1 Of 8), Other |
Other |
2000 |
0.22 |
Functional ≤ 10μM
|
|
Z50466-4-O |
Trypanosoma Cruzi (cluster #4 Of 8), Other |
Other |
9800 |
0.19 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
37 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.77 |
12.55 |
-49.34 |
1 |
8 |
1 |
70 |
532.448 |
7 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
4369693
-
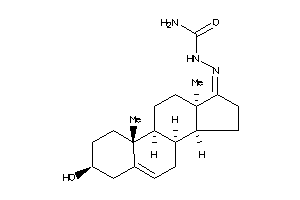
-
4369694
-
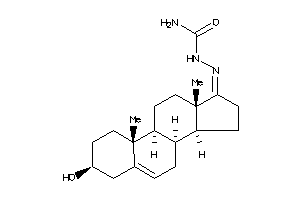
-
13467051
-
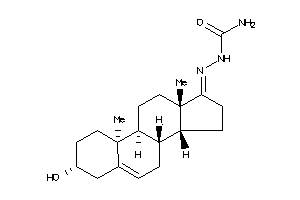
-
13467053
-
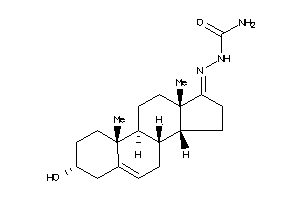
-
13467055
-
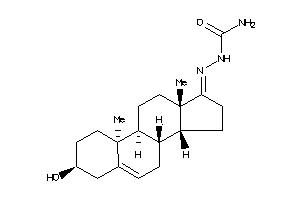
Draw
Identity
99%
90%
80%
70%
Popular Name:
2-(3-hydroxy-10,13-dimethyl-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yliden)hydrazine-1-carboxamide
2-(3-hydroxy-10,13-dimethyl-2,3,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-2-E |
Cytochrome P450 17A1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
178 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
3.82 |
-8.3 |
4 |
5 |
0 |
88 |
345.487 |
1 |
↓
|
|