|
|
Analogs
-
39241039
-
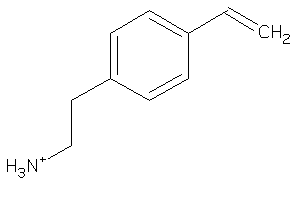
Draw
Identity
99%
90%
80%
70%
Vendors
And 41 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TAAR1-2-E |
Trace Amine-associated Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4380 |
0.75 |
Functional ≤ 10μM
|
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.76 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5300 |
0.74 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
3.99 |
-44.19 |
3 |
1 |
1 |
28 |
136.218 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 46 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3280 |
0.96 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3920 |
0.95 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.73 |
4.22 |
-6.49 |
0 |
1 |
0 |
17 |
106.124 |
1 |
↓
|
|
|
|
Analogs
-
34426560
-
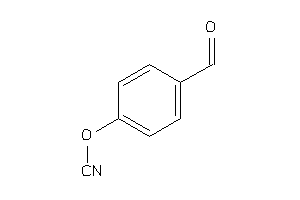
Draw
Identity
99%
90%
80%
70%
Vendors
And 55 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5230 |
0.74 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5150 |
0.74 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
3.53 |
-7.83 |
0 |
2 |
0 |
26 |
136.15 |
2 |
↓
|
|
|
|
Analogs
-
1729595
-
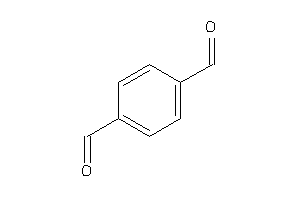
Draw
Identity
99%
90%
80%
70%
Vendors
And 59 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4680 |
0.83 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4880 |
0.83 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
1.93 |
-6.42 |
0 |
1 |
0 |
17 |
120.151 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
1.05 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
0.91 |
-1.48 |
0 |
0 |
0 |
0 |
147.004 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP1A2-2-E |
Cytochrome P450 1A2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2300 |
0.66 |
ADME/T ≤ 10μM
|
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5500 |
0.61 |
ADME/T ≤ 10μM
|
|
CP2A6-5-E |
Cytochrome P450 2A6 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
2500 |
0.65 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.41 |
7.61 |
-2.8 |
0 |
0 |
0 |
0 |
197.064 |
0 |
↓
|
|
|
|
Analogs
-
33690081
-
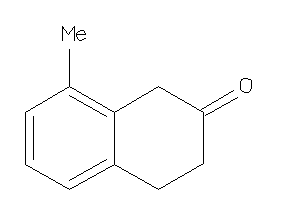
-
36683743
-
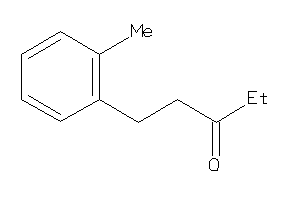
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9705 |
0.64 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
6.04 |
-5.48 |
0 |
1 |
0 |
17 |
146.189 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7300 |
0.65 |
ADME/T ≤ 10μM
|
|
CP2A6-5-E |
Cytochrome P450 2A6 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
670 |
0.79 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
6.84 |
-3.85 |
0 |
0 |
0 |
0 |
146.164 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 44 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.71 |
ADME/T ≤ 10μM
|
|
CP2A6-5-E |
Cytochrome P450 2A6 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
550 |
0.80 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
7.37 |
-3.15 |
0 |
0 |
0 |
0 |
207.07 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.67 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
1.96 |
-3.58 |
0 |
0 |
0 |
0 |
156.228 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.68 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1700 |
0.67 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
8.08 |
-3.61 |
0 |
0 |
0 |
0 |
156.228 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 41 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.99 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
-0.32 |
-1.38 |
0 |
0 |
0 |
0 |
235.906 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.66 |
ADME/T ≤ 10μM
|
|
CP2A6-5-E |
Cytochrome P450 2A6 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
2500 |
0.65 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.41 |
7.61 |
-2.94 |
0 |
0 |
0 |
0 |
197.064 |
0 |
↓
|
|
|
|
Analogs
-
39384787
-
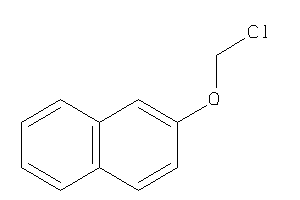
Draw
Identity
99%
90%
80%
70%
Vendors
And 33 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4603 |
0.62 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
1.34 |
-5.07 |
0 |
1 |
0 |
9 |
158.2 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 30 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.66 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2400 |
0.72 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
7.42 |
-3.56 |
0 |
0 |
0 |
0 |
142.201 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP1A2-2-E |
Cytochrome P450 1A2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7900 |
0.60 |
ADME/T ≤ 10μM
|
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9908 |
0.58 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8300 |
0.59 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
7.95 |
-3.78 |
0 |
0 |
0 |
0 |
156.228 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4800 |
0.62 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.04 |
8.19 |
-3.34 |
0 |
0 |
0 |
0 |
156.228 |
1 |
↓
|
|
|
|
Analogs
-
39250338
-
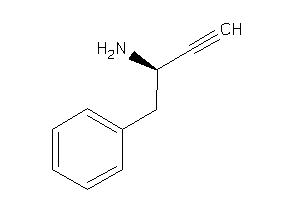
-
39250339
-
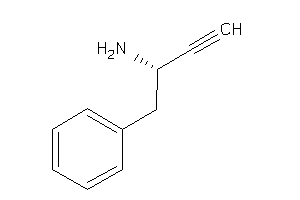
Draw
Identity
99%
90%
80%
70%
Vendors
-
- API0000117
- CCH0003609
- API0002279
-
-
And 12 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
4.01 |
-39.14 |
3 |
1 |
1 |
28 |
136.218 |
2 |
↓
|
|
|
|
Analogs
-
39250338
-

-
39250339
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7660 |
0.72 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7660 |
0.72 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7660 |
0.72 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7660 |
0.72 |
Binding ≤ 10μM
|
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4900 |
0.74 |
Binding ≤ 10μM |
|
TAAR1-2-E |
Trace Amine-associated Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
920 |
0.85 |
Functional ≤ 10μM
|
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2850 |
0.78 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3500 |
0.76 |
ADME/T ≤ 10μM
|
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
490 |
0.88 |
Functional ≤ 10μM
|
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
800 |
0.85 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
4 |
-39.2 |
3 |
1 |
1 |
28 |
136.218 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 60 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TAAR1-2-E |
Trace Amine-associated Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
129 |
1.07 |
Functional ≤ 10μM
|
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.86 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4410 |
0.83 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.92 |
3.31 |
-44.08 |
3 |
1 |
1 |
28 |
122.191 |
2 |
↓
|
|